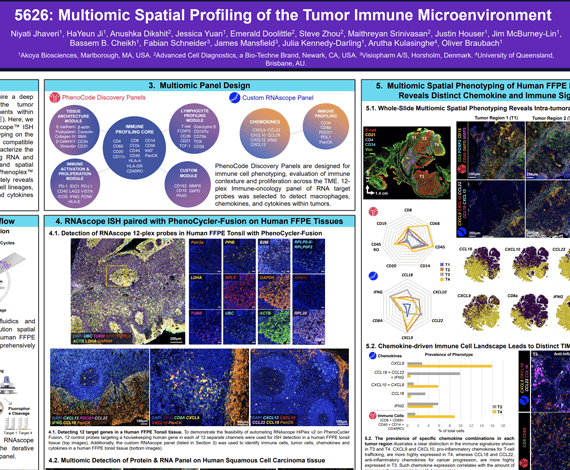
It has been well established that the tumor microenvironment (TME), which comprises cancer cells, stromal cells, and surrounding extracellular matrix, plays a critical role in cancer development, progression, and control. The immunological components within tumors, known as the tumor immune microenvironment (TiME), have also been implicated in tumor development, recurrence, and metastasis. Effective strategies for cancer immunotherapies will require a deep understanding of the factors that shape both the TME and TiME. Here, we describe a spatial multiomics approach that utilizes RNAscope™ ISH technology paired with high-plex whole-slide spatial phenotyping with the PhenoCycler™-Fusion platform. This two-step approach is compatible with human FFPE tissues and enables researchers to characterize the spatial biology of the TiME more accurately by detecting RNA and protein markers on serial sections. The resulting multiomic data more accurately reveal the interplay between TME and TiME by giving insight into cell lineages, surrounding structures, as well as secreted chemokines and cytokines that exist within the TME ecosystem.
Niyati Jhaveri1, HaYeun Ji1, Anushka Dikshit2, Jessica Yuan1, Emerald Doolittle2, Steve Zhou2, Maithreyan Srinivasan2, Bassem B. Cheikh1, Fabian Schneider3, James Mansfield3, Julia Kennedy-Darling1, Oliver Braubach1
- Akoya Biosciences, Menlo Park, CA
- Advanced Cell Diagnostics, a Bio-Techne Brand, Newark, CA
- Visiopharm A/S, Horsholm, Denmark

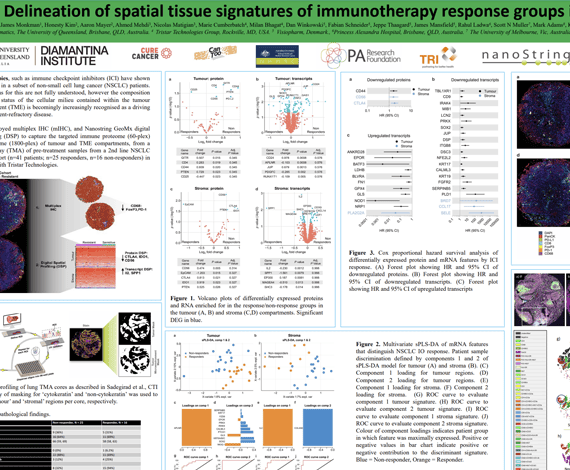
The growth in cancer immunotherapy agents requires an understanding of the immune contexture of the tumor microenvironment (TME). One way to understand immune contexture is to use multiplex staining, imaging, and analysis to obtain multi-marker phenotypes of specific cells and analyze their biodistribution in the TME. Imaging Mass Cytometry™ (IMC) is the method of choice for single-step staining and highplex imaging of FFPE tissues. FFPE tissue is autofluorescent, which limits the utility of immunofluorescence methods, particularly when done without amplification. Lung and colorectal tissue (and bone, skin, etc) are highly autofluorescence, and therefore are a good target for IMC imaging, which has no autofluorescence issues. However, developments in analysis software with a single-package workflow for highplex imagery have not kept pace. We present here a comprehensive workflow in the Oncotopix® Discovery platform designed specifically for highplex IMC image analysis, covering tissue segmentation, cell segmentation based on IMC DNA images, cellular phenotyping, and spatial analyses.
Fabian Schneider1, Brenna O’Neill1, Rasmus A. Lyngby1, Rasmus N. Sørensen1, Andreas Hussing1, Alessandro S. Massaro1, Andrew Quong2, Smriti Kala2, Sam Lim2, Clinton Hupple2, Nina Lane2, Michelle Macpherson2, Jeppe Thagaard1, Johan Doré Hansen1, James Mansfield1
- Visiopharm, Horsholm, Denmark
- Standard BioTools, South San Francisco, CA, USA


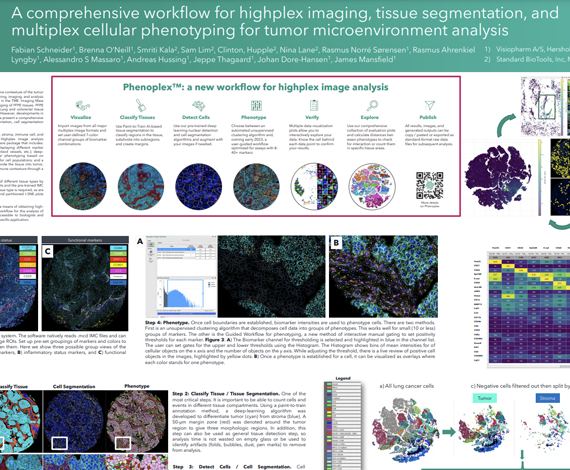
Imaging Mass Cytometry™ (IMC™) is a powerful tool for the study of complex cellular interactions in the tumor microenvironment (TME) and in the discovery of biomarkers that can predict disease outcome or response to therapy. The Hyperion™ Imaging System (Standard BioTools™) utilizes CyTOF® technology to simultaneously assess 40-plus protein markers at subcellular resolution without spectral overlap or background autofluorescence, thus providing unprecedented insight into the organization and function of the TME. However, despite the advances in staining and imaging methods, developments in analysis software have not kept pace, as we lack a complete, user-defined workflow in a single software package for analysis of high-plex imaging data. Here we present a comprehensive workflow using the Visiopharm® Phenoplex™ platform, designed specifically for high-plex IMC image analysis.
Smriti Kala1, Brenna O’Neill2, Rasmus Lyngby2, Rasmus Sorensen2, Andrew Quong1, Sam Lim1, Clinton Hupple1, Nina Lane1, Jeppe Thagaard2, Johan Dore Hansen2, James Mansfield2, Fabian Schneider2, Michelle Macpherson1
- Standard BioTools Canada Inc., Markham, Ontario, Canada
- Visiopharm A/S, Hørsholm, Denmark
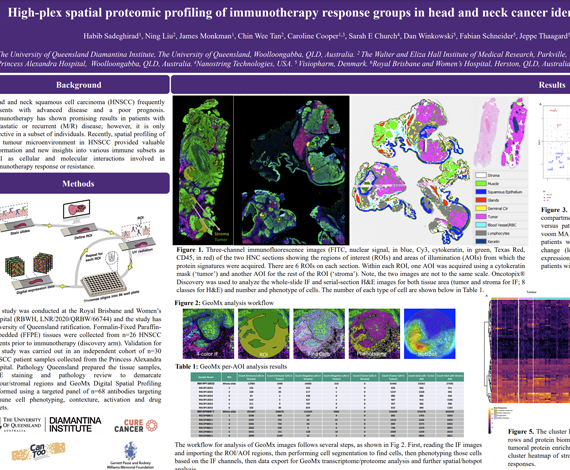

Head and neck squamous cell carcinoma (HNSCC) frequently presents with advanced disease and a poor prognosis. Immunotherapy has shown promising results in patients with metastatic or recurrent (M/R) disease; however, it is only effective in a subset of individuals. Recently, spatial profiling of the tumour microenvironment in HNSCC provided valuable information and new insights into various immune subsets as well as cellular and molecular interactions involved in
immunotherapy response or resistance.
Habib Sadeghirad1, Ning Liu2, James Monkman1, Chin Wee Tan2, Caroline Cooper1,3, Sarah E Church4, Dan Winkowski5, Fabian Schneider5, Jeppe Thaagard5, James Mansfield5, Ken O’Byrne3, Melissa Davis2, Brett Hughes6, Arutha Kulasinghe1
-
- The University of Queensland Diamantina Institute, The University of Queensland, Woolloongabba, QLD, Australia.
- The Walter and Eliza Hall Institute of Medical Research, Parkville, VIC, Australia.
- Princess Alexandra Hospital, Woolloongabba, QLD, Australia.
- Nanostring Technologies, USA.
- Visiopharm, Denmark.
- Royal Brisbane and Women’s Hospital, Herston, QLD, Australia
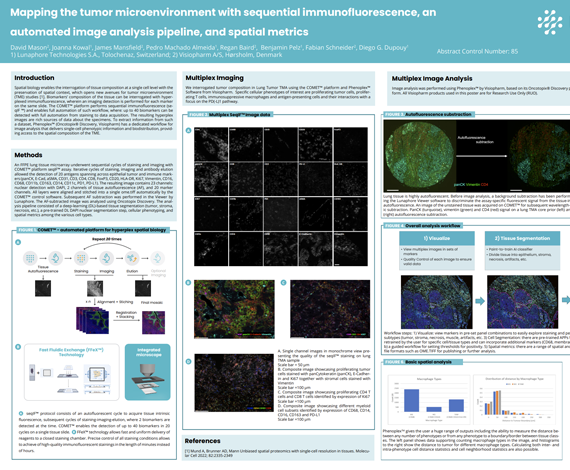

Spatial biology enables the interrogation of tissue composition at a single cell level with the preservation of spatial context, which opens new avenues for tumor microenvironment (TME) studies [1]. Biomarkers’ composition of the tissue can be interrogated with hyperplexed immunofluorescence, wherein an imaging detection is performed for each marker on the same slide. The COMET™ platform performs sequential immunofluorescence (seqIF ™) and enables full automation of such workflow, where: up to 40 biomarkers can be detected with full automation from staining to data acquisition. The resulting hyperplex images are rich sources of data about the specimens. To extract information from such a dataset, Phenoplex™ (Oncotopix® Discovery, Visiopharm) has a dedicated workflow for image analysis that delivers single-cell phenotypic information and biodistribution, providing access to the spatial composition of the TME.
David Mason2, Joanna Kowal1, James Mansfield2, Pedro Machado Almeida1, Regan Baird2, Benjamin Pelz1, Fabian Schneider2, Diego G. Dupouy1
- Lunaphore Technologies S.A., Tolochenaz, Switzerland
- Visiopharm A/S, Hørsholm, Denmark
Immunotherapies, such as immune checkpoint inhibitors (ICI) have shown durable benefit in a subset of non-small cell lung cancer (NSCLC) patients. The mechanisms for this are not fully understood, however the composition and activation status of the cellular milieu contained within the tumour microenvironment (TME) is becomingly increasingly recognised as a driving factor in treatment-refractory disease.
Here, we employed multiplex IHC (mIHC), and Nanostring GeoMx digital spatial profiling (DSP) to capture the targeted immune proteome (60-plex) and transcriptome (1800-plex) of tumour and TME compartments, from a tissue microarray (TMA) of pre-treatment samples from a 2nd line NSCLCICI-treated cohort (n=41 patients; n=25 responders, n=16 non-responders) in collaboration with Tristar Technologies.
Arutha Kulasinghe1, James Monkman1, Honesty Kim2, Aaron Mayer2, Ahmed Mehdi3, Nicolas Matigian3, Marie Cumberbatch4, Milan Bhagat4, Dan Winkowski5, Fabian Schneider5, Jeppe Thaagard5, James Mansfield5, Rahul Ladwa6, Scott N Muller7, Mark Adams8, Ken O’Byrne8
-
- University of Queensland Diamantina Institute, The University of Queensland, Brisbane, QLD, Australia
- Enable Medicine, Menlo Park, CA, USA
- Qfab Bioinformatics, The University of Queensland, Brisbane, QLD, Australia
- Tristar Technologies Group, Rockville, MD, USA
- Visiopharm, Denmark
- Princess Alexandra Hospital, Brisbane, QLD, Australia
- The University of Melbourne, Vic, Australia
- Queensland University of Technology, Brisbane, QLD, Australia