The advancement of digital pathology has led to revolutionary improvements in patient care and medical research. The cellular microenvironment is complex, but the ability of researchers to interrogate it has improved with the development of technologies such as Imaging Mass CytometryTM (IMCTM, Fluidigm) and high-plex fluorescent staining panels (e.g. Akoya or Ultivue). However, increases in the amount and complexity of the data that these technologies yield has brought on a new challenge: developing methods capable of detecting meaningful differences in the datasets.
Introduction to multispectral imaging and analysis
Dr. Heather Stevenson is Associate Professor at the University of Texas Medical Branch and Director of Transplantation Pathology. Her primary research focus is hepatic immunology and how dysregulation of the immune response leads to fibrosis development. She is researching how hepatic macrophage phenotypes determine immune activation and reaction to liver injury. In her latest study, she investigated the phenotypes of intrahepatic macrophages in patients with different types of chronic liver disease.
To obtain a full picture of the microenvironment and to distinguish multiple cell populations, multiple specific biomarkers are needed. To safely assess those biomarkers, various new techniques have been developed, like flow or mass cytometry. Typically those methods dissolve the local context, so that localizations and relations of the cell populations cannot be measured and valuable information is lost. Flow cytometry is unable to visualize multiple antigens in the context of hepatic architecture, and also requires fresh human tissue. It cannot be used for FFPE tissue, which is often more readily available. Single-cell RNA sequencing and mass cytometry can detect multiple markers on intrahepatic macrophages, which is an improvement compared to flow cytometry, but neither can preserve the hepatic architecture or detect locations of the identified cell populations.
Additionally, macrophages are difficult to work with in cell culture, as they can change phenotype when cultured and are not easily isolated from human liver tissue. Mouse models cannot be used for phenotyping intrahepatic macrophages, as they do not adequately replicate the long-term chronic diseases that commonly affect humans, including most liver diseases.
The analyses that most closely replicate the in vivo hepatic microenvironment are in situ methods, rendering multispectral imaging of the tissue slide the method of choice for this type of investigation. Multiplex image analysis is commonly used in immunology research, very prominently in immune-oncology, and also in the investigation of immune cell types in the liver. It also allows to maximize the information collected from a single tissue slide. This is important, since the liver biopsies are being collected in an invasive procedure and are limited, so more information may allow to personalize treatment in the future depending on cellular phenotypes present.
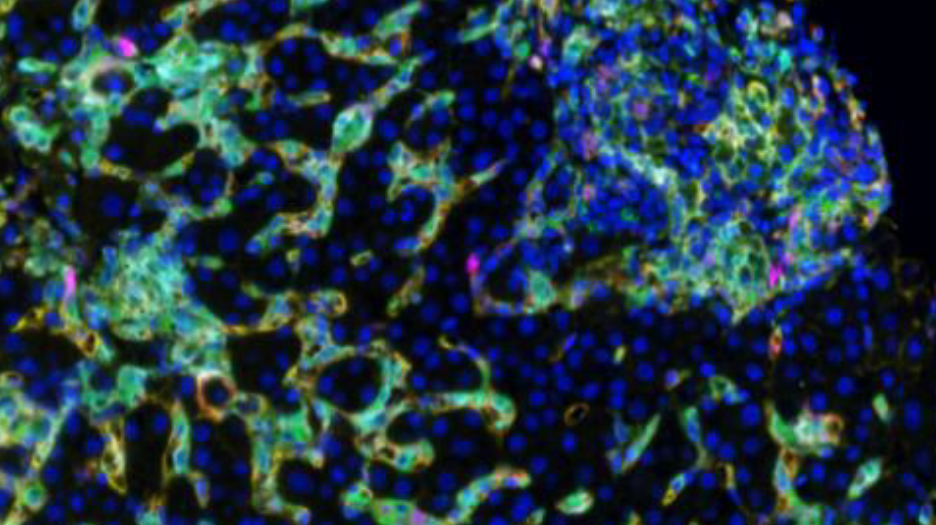
In a recent project, Dr. Stevenson used spectral imaging microscopy (Vectra 3, Akoya Biosystems) to phenotype intrahepatic macrophages in patients with different chronic diseases of the liver, including chronic hepatitis C (HCV), nonalcoholic steatohepatitis (NASH) and autoimmune hepatitis (AIH). Dr. Stevenson and her team analyzed their image data using Visiopharm’s multiplex phenotyping module, which provides efficient and reproducible analyses of high-dimensional multiplexed images.
The technical challenges of working with liver tissue
The first challenge of the study was the high background autofluorescence that liver tissue tends to show due to the high amount of fluorophores. This usually requires specialized equipment to perform spectral unmixing (done by the Vectra System), which removes background autofluorescence from the meaningful data. Following application of the multiplex staining protocols, a large volume of data is generated, especially if researchers are using batch analysis with multiple patients per group. The Visiopharm software robustly copes with the IF images obtained from the Vectra System and facilitates these big data analyses increasing efficiency and reproducibility.
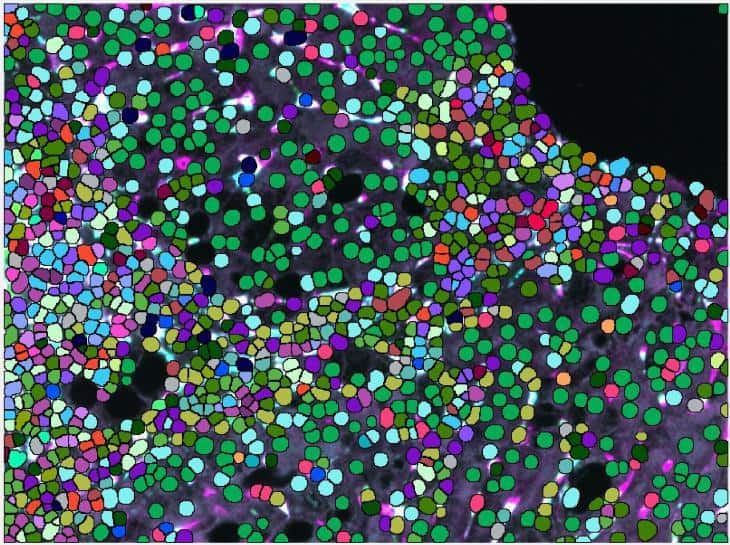
A second challenge presented by this liver tissue study was the difficulty of macrophage segmentation, which can be problematic due to the morphometry of macrophages. It is especially demanding in liver tissue since macrophages are not the primary cell type and their boundaries are often obscured when they are among hepatocytes and epithelial cells.
Very important to the phenotype assessment was also to use an unsupervised approach to deliver the spectrum of phenotypes for all investigated chronic liver diseases and be able to compare those. Using less advanced image mining tool, this step can easily introduce bias to the data and tends to be rather tedious, keeping in mind, that there were 32 possible combinations of macrophage subpopulations to determine per patient and compare between the groups.
Visiopharm’s strategies to overcome liver tissue challenges
Visiopharm’s pretrained AI analysis tools can accurately segment macrophages without additional training of the algorithm, which is an advantage compared to other deep learning platforms that need to be trained from scratch on large annotated datasets. This can take time and also create bias, depending on the training dataset that is used and especially if unsufficient training data is available. The VIS AI tools were able to easily identify the cells for analysis without the need for further training.
The phenotype module of Visiopharm’s software provides the exact framework for analyzing any dataset, by offering the possibility to take an agnostic approach and allowing the data itself to dictate the downstream results, rather than having them be influenced by a priori expectations. “We were quite surprised at the accuracy and precision of the automated phenotyping and the simplicity in using the software,” Dr. Stevenson says of Visiopharm’s phenotyping module. “One of the really interesting findings that is consistent with our overall thoughts is that CD68 and CD163 are not found on the same cells, despite being fairly interchangeable in most of the macrophage literature.” This suggests, that the Visiopharm software had detected differences in cell phenotypes that were not previously known to researchers, which is revolutionary not only for researchers studying liver diseases, but also for all scientists working in the field of immunology.
Visiopharm’s software also provides advanced visualizations to investigate, compare and confirm the results of the analysis. Plots of the dimensionality-reduced data by the t-distributed stochastic neighbor embedding (tSNE) algorithm showed clear differences between the groups of Dr. Stevenson’s study. The data points from each individual case were mapped to the same coordinates, making comparisons between the patients with different liver diseases simple and efficient.
Phenotype color coding was matched between the tSNE plots and the image annotations, which allowed the images to be easily referenced. The phenotypic profile plots showed relative expression levels of different macrophage phenotypes across patient groups in a single simple graphical representation. These software features greatly facilitate what is typically an extremely complex and time-consuming analysis.
Future directions following the initial study
The results of this study allowed researchers in Dr. Stevenson’s lab to expand their panel to other immune cell types, as well as more specific immune cell subtypes. “We can now probe the liver for specific macrophage subsets in addition to other cell types to better understand mechanisms of liver disease and disease signatures,” explains Dr. Stevenson.
Spectral imaging microscopy followed by Visiopharm’s software image analysis is a novel method of phenotyping intrahepatic macrophages in patients with different liver pathologies and has the potential to change the current understanding of intrahepatic macrophages, as well as the way that patient liver biopsies are evaluated in the future.
More information is available by attending this free webinar.
The study is published in Hepatology Communications.
 Visiopharm
Categories: Blog
Visiopharm
Categories: Blog
