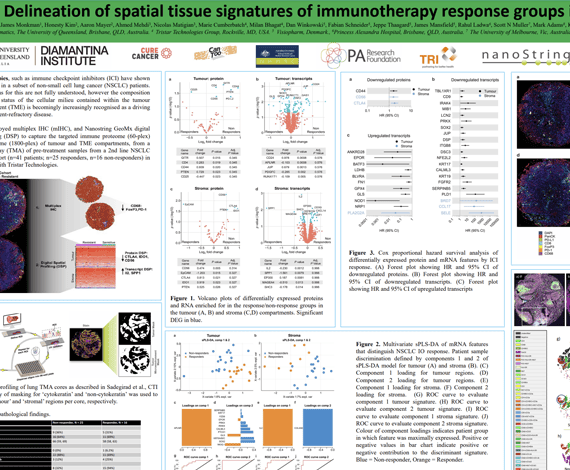
Immunotherapies, such as immune checkpoint inhibitors (ICI) have shown durable benefit in a subset of non-small cell lung cancer (NSCLC) patients. The mechanisms for this are not fully understood, however the composition and activation status of the cellular milieu contained within the tumour microenvironment (TME) is becomingly increasingly recognised as a driving factor in treatment-refractory disease.
Here, we employed multiplex IHC (mIHC), and Nanostring GeoMx digital spatial profiling (DSP) to capture the targeted immune proteome (60-plex) and transcriptome (1800-plex) of tumour and TME compartments, from a tissue microarray (TMA) of pre-treatment samples from a 2nd line NSCLCICI-treated cohort (n=41 patients; n=25 responders, n=16 non-responders) in collaboration with Tristar Technologies.
Arutha Kulasinghe1, James Monkman1, Honesty Kim2, Aaron Mayer2, Ahmed Mehdi3, Nicolas Matigian3, Marie Cumberbatch4, Milan Bhagat4, Dan Winkowski5, Fabian Schneider5, Jeppe Thaagard5, James Mansfield5, Rahul Ladwa6, Scott N Muller7, Mark Adams8, Ken O’Byrne8
-
- University of Queensland Diamantina Institute, The University of Queensland, Brisbane, QLD, Australia
- Enable Medicine, Menlo Park, CA, USA
- Qfab Bioinformatics, The University of Queensland, Brisbane, QLD, Australia
- Tristar Technologies Group, Rockville, MD, USA
- Visiopharm, Denmark
- Princess Alexandra Hospital, Brisbane, QLD, Australia
- The University of Melbourne, Vic, Australia
- Queensland University of Technology, Brisbane, QLD, Australia