The ability to image tissue microenvironments (TME) at high-plex over an entire slide without requiring multiple staining steps allows for unprecedented insights into tissue architecture and the molecular mechanisms of immune and disease processes. Here we investigate a whole slide tissue section of high-grade colon adenocarcinoma, a hot tumor that contains prominent clusters of PD-L1 expression scattered throughout, using single-step high-plex staining and imaging at single-cell resolution followed by the analysis of single-cell phenotypes, tissue segmentation and spatial proximity and nearest neighbor analysis.
Fabian Schneider1, James Mansfield1, Edward Lo2, Tad George2, Joshua Nordberg2
- Visiopharm A/S, Horsholm, Denmark
- RareCyte, Seattle, WA
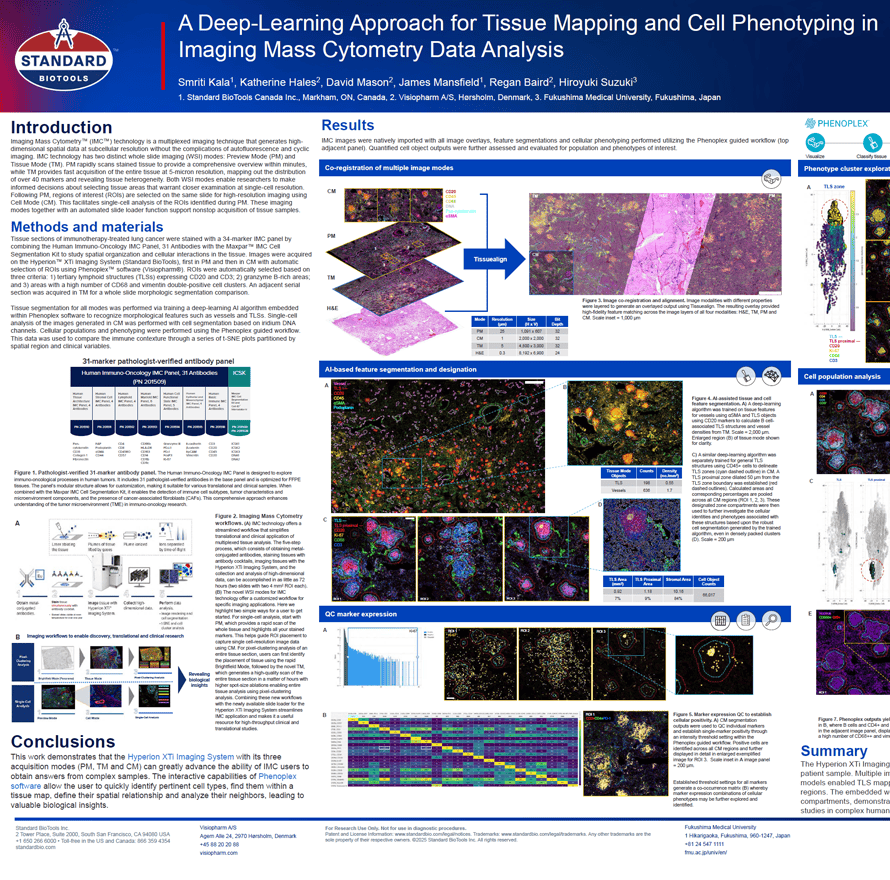
Imaging Mass Cytometry (IMC ) technology is a multiplexed imaging technique that generates high-dimensional spatial data at subcellular resolution without the complications of autofluorescence and cyclic imaging. IMC technology has two distinct whole slide imaging (WSI) modes: Preview Mode (PM) and Tissue Mode (TM). PM rapidly scans stained tissue to provide a comprehensive overview within minutes, while TM provides fast acquisition of the entire tissue at 5-micron resolution, mapping out the distribution of over 40 markers and revealing tissue heterogeneity. Both WSI modes enable researchers to make informed decisions about selecting tissue areas that warrant closer examination at single-cell resolution. Following PM, regions of interest (ROIs) are selected on the same slide for high-resolution imaging using Cell Mode (CM). This facilitates single-cell analysis of the ROIs identified during PM. These imaging modes together with an automated slide loader function support nonstop acquisition of tissue samples.
Smriti Kala1, Katherine Hales2, David Mason2, James Mansfield1, Regan Baird2, Hiroyuki Suzuki3
- Standard BioTools Canada Inc., Markham, ON, Canada
- Visiopharm A/S, Hørsholm, Denmark
- Fukushima Medical University, Fukushima, Japan
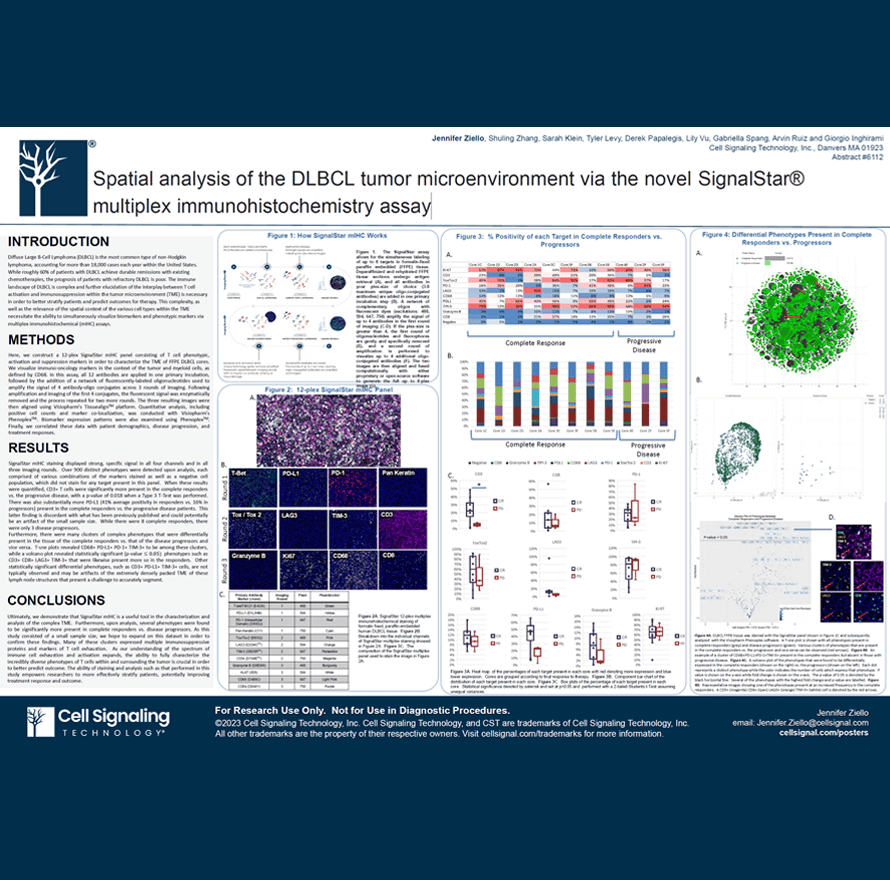
Diffuse Large B-Cell Lymphoma (DLBCL) is the most common type of non-Hodgkin lymphoma, accounting for more than 18,000 cases each year within the United States. While roughly 60% of patients with DLBCL achieve durable remissions with existing chemotherapies, the prognosis of patients with refractory DLBCL is poor. The immune landscape of DLBCL is complex and further elucidation of the interplay between T cell activation and immunosuppression within the tumor microenvironment (TME) is necessary in order to better stratify patients and predict outcomes for therapy. This complexity, as well as the relevance of the spatial context of the various cell types within the TME necessitate the ability to simultaneously visualize biomarkers and phenotypic markers via multiplex immunohistochemical (mIHC) assays.
Jennifer Ziello, Shuling Zhang, Sarah Klein, Tyler Levy, Derek Papalegis, Lily Vu, Gabriella Spang, Arvin Ruiz and Giorgio Inghirami
Cell Signaling Technology, Inc., Danvers MA 01923
A novel global phenotyping method using artificial intelligence to overcome the cell segmentation bottleneck
A novel global phenotyping method using artificial intelligence to overcome the cell segmentation bottleneck


Multiplex methods for the detection of protein expression generate extremely data-rich images of intact tissue sections. These images are invaluable for the quantification and analysis of complex biology and have huge potential for biomarker development. However, their interpretation presents a considerable data analysis challenge which limits their utility, especially in clinical workflows.
The key task from these images is often cellular phenotyping – e.g. the identification of immunophenotypes, or the separation cells with compartment-specific protein expression from others. Most current automated methods depend upon a cellular segmentation step to support this, and segmentation has become an area of intensive research in AI (Artificial Intelligence), but the task is very difficult and no universally accepted method has been identified.
Therefore, methods of cellular phenotyping which operate without the need for segmentation would be very valuable. To meet this challenge, we have developed an AI cellular phenotyping method which does not require segmentation at all.
We use the U-Net backbone within the Visiopharm deep learning module to train our algorithm using expert human annotations of entire cellular regions. Crucially, we need only train for a single example of each compartmental stain (nuclear/cytoplasmic/membranous), and the algorithm then assigns class identities to cells without the need for segmentation of nuclei cytoplasm or membranes, using regional information in a manner analogous to a human expert, who may not need to be confident about cellular boundaries to accurately phenotype a cell.
The method is comparable in accuracy to segmentation-based methods, is highly transferrable, and represents a highly novel approach to quantitative image analysis with the potential to radically alter the way that we analyze these data-rich images.

Rachel Pennie, MSc
Rachel Pennie MSc is a translational histologist in the deep phenotyping advanced technology facility within the Le Quesne Lab at the CRUK Scotland Institute. She is an expert in the development and application of Visiopharm deep learning methods to multiplex and high plex histopathological images.
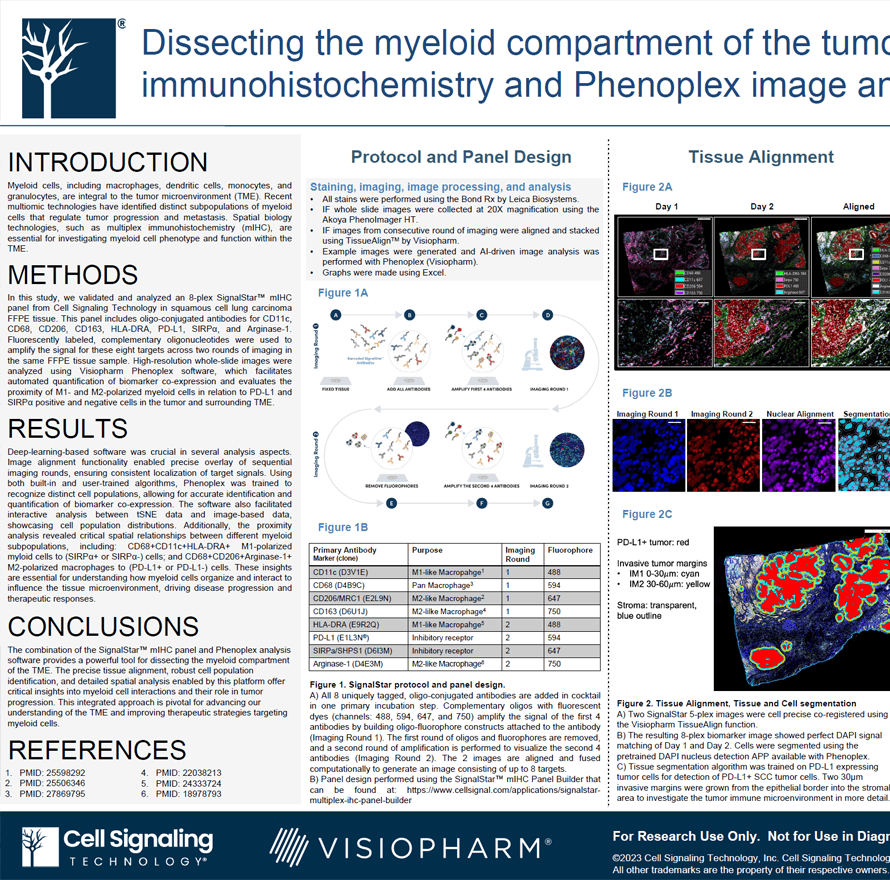

Myeloid cells, including macrophages, dendritic cells, monocytes, and granulocytes, are integral to the tumor microenvironment (TME). Recent multiomic technologies have identified distinct subpopulations of myeloid cells that regulate tumor progression and metastasis. Spatial biology technologies, such as multiplex immunohistochemistry (mIHC), are essential for investigating myeloid cell phenotype and function within the TME.
Sarah R Klein1, Lily Vu1, James Mansfield2, Fabian Schneider2
- Cell Signaling Technology, Inc., Danvers MA, USA
- Visiopharm A/S, Horsholm, Denmark
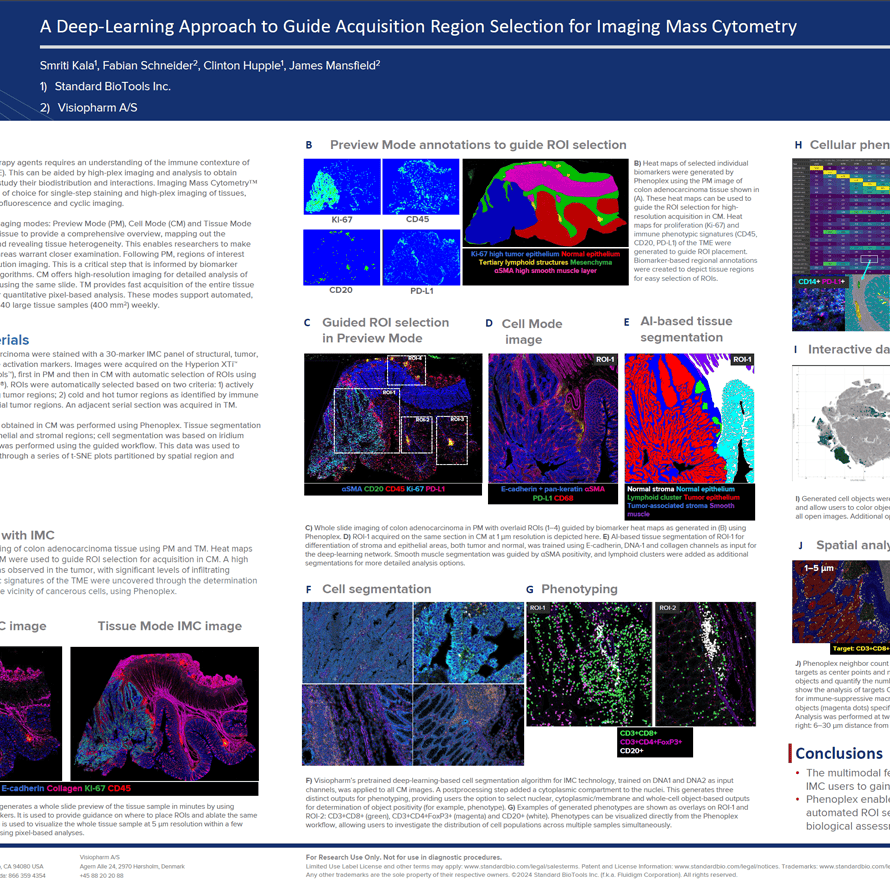
The growth in cancer immunotherapy agents requires an understanding of the immune contexture of the tumor microenvironment (TME). This can be aided by high-plex imaging and analysis to obtain phenotypes of specific cells and study their biodistribution and interactions. Imaging Mass Cytometry (IMC) is the method of choice for single-step staining and high-plex imaging of tissues, avoiding the complications of autofluorescence and cyclic imaging.
IMC has expanded its capabilities with three distinct imaging modes: Preview, Cell, and Tissue. The Preview Mode is a rapid scanning system that captures a comprehensive overview of the stained tissue, mapping out the distribution of over 40 markers and revealing tissue heterogeneity. This enables researchers to make informed decisions about which areas warrant closer examination on the same. Building on this, Cell Mode offers high-resolution imaging for detailed analysis of the Regions of Interest (ROIs) identified during Preview, all using the same slide. Tissue Mode complements these by providing a fast acquisition of the entire tissue at a lower resolution, which is optimal for quantitative pixel-based analysis of tissue biology. These modes support automated, continuous imaging of more than 40 large tissue samples (400 mm2) weekly. Following Preview Mode, the selection of ROIs for high-resolution imaging is a critical step, enhanced by automated AI algorithms to ensure it is informed by biomarker expression.
Smriti Kala1, Fabian Schneider2, Clinton Hupple1, James Mansfield2
- Standard BioTools Inc.
- Visiopharm A/S