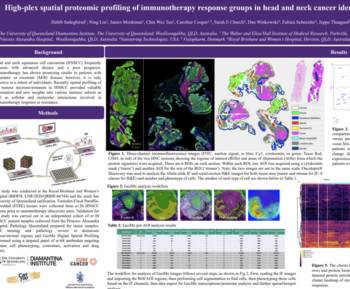
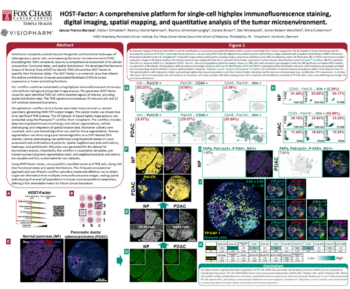
Head and neck cancer (HNC) is a heterogeneous group of malignancies that arise from the mucosal surfaces of the upper aerodigestive tract. The tumor microenvironment (TME) of HNC is characterized by the presence of immune cells, stromal cells, and extracellular matrix components. A key feature of the TME is hypoxia, which promotes tumor growth, invasion, and metastasis by altering the expression of genes involved in angiogenesis, cell survival, and metabolism. Understanding the complex interplay between hypoxia and immune infiltrates in the TME of HNC is crucial for the development of novel therapeutic strategies for the treatment of this disease. Whole transcriptome analysis by digital spatial profiling is an excellent method of probing the TME, but assessing large cohorts can be time consuming. Automating a profiling workflow to reduce hands-on time and region of interest (ROI) selection bias will enable exploration of large cohorts to identify mechanisms of action, potential drug targets, and biomarkers.
Jeni Caldara1, Sarah E. Church2, Regan Baird1, David Mason1, Kyla Teplitz2, Kristina Sorg2, Megan Vandenberg2, James Mansfield1, Kelly Hunter3, Joana Campos3, Hisham Mehanna4, Jill Brooks4
- Visiopharm, Horsholm, Denmark
- NanoString Technologies, Seattle, WA, USA
- Propath, Hereford, UK
- Institute of Head and Neck Studies and Education, U Birmingham, Birmingham, UK