
Q&A: OracleBio’s journey with Visiopharm’s software
In today’s blog, dive into the engaging Q&A session with OracleBio, a pioneering CRO in quantitative digital pathology, as they discuss their journey with Visiopharm’s software.
The interview with Gabriel Reines March (R&D Project Manager at OracleBio) and Karen McClymont (Image Analysis Project Manager at OracleBio) offers an insight into how OracleBio leverages cutting-edge image analysis and AI technologies to enhance research and development in pharma and biotech globally. From innovative algorithm development to their transition to a cloud-based infrastructure, discover the pivotal role Visiopharm plays in OracleBio’s work, especially in the fields of oncology and immuno-oncology.

Gabriel Reines March
R&D Project Manager
With a PhD in Biomedical Image Processing and a background in Electrical Engineering, Gabriel is the OracleBio R&D team lead. He oversees and manages the group’s project pipeline and ensures that the company stays at the bleeding edge of the industry.
Gabriel also manages OracleBio’s involvement in the INCISE project – a collaborative effort between industry, academia and the UK’s National Health Service.

Karen McClymont
Image Analysis Project Manager
With a PhD in Biochemistry and her active involvement and support in the analysis of some of OracleBio’s most complex studies, Karen plays a key role leading OracleBio’s image analysis projects and overseeing the team’s continued development.
Visiopharm: Can you tell me a bit about OracleBio and which services you are offering to your customers?
OracleBio: OracleBio is a leading Contract Research Organization (CRO) that provides quantitative digital pathology services to Pharma and Biotech globally. We help our customers by generating accurate and robust data from their histology images, enabling confident interpretation and efficient decision making for their R&D projects.
As well as experienced image analysts, our team is also made up of clinical pathologists, software engineers, and Pharma scientists. This broad expertise base allows us to advise our clients on the best histology staining and analysis approach to ensure relevant image analysis data can be generated in the most optimal manner.
We utilise commercially available image analysis software like Visiopharm and support all stages of the R&D pipeline, from target discovery to clinical trials, with a distinct edge in oncology, immuno-oncology, and fibrosis. Our expertise extends to a diverse range of histological techniques, such as H&E, Immunohistochemistry, Multiplex Immunofluorescence, in situ Hybridization, and Imaging Mass Cytometry. The power and versatility of the software, with the addition of deep learning capabilities, is well-suited to the innovative way OracleBio approaches algorithm development for the multitude of staining profiles that we encounter.
Important to the efficient delivery of data is our image analysis workflow, hosted on our purpose-built, state-of-the-art Amazon Web Services (AWS) cloud that is designed to enable quantitative digital pathology workflows at speed and scale. What’s more, in order to facilitate client engagement and collaboration throughout the study’s lifecycle, we have created our own Visiopharm-powered remote image viewing portal, OBserver, where clients can log on to generate annotations and review analysis overlays.
The power and versatility of the software, with the addition of deep learning capabilities, is well-suited to the innovative way OracleBio approaches algorithm development for the multitude of staining profiles that we encounter.

Visiopharm: Since when have you been using Visiopharm and what led you to Visiopharm originally?
OracleBio: As a Digital Pathology CRO, our primary focus is to generate robust data from our clients’ images. Having a powerful and flexible image analysis software such as Visiopharm provides us with options for innovative solutions to the more challenging image analysis problems. We integrated Visiopharm into our roster of image analysis tools back in 2016. Some of the software features that attracted us were:
- the inclusion of different Deep Learning neural networks to train and deploy tissue classifier and cell segmentation apps;
- the availability of a comprehensive collection of image post-processing steps, useful for customizing apps to suit specific study requirements;
- the flexibility around app development for building bespoke image analysis workflows.
For these reasons, Visiopharm quickly became an integral part of our image analysis workflow.
Visiopharm: Can you describe the process of implementing Visiopharm and how your experience has been working with our team?
OracleBio: When we first introduced Visiopharm into OracleBio’s image analysis workflow, the software was run locally on our on-premise server, which meant we only had access to two user licenses and finite hardware resources. Although this model suited our needs at the time, the limitations of such a rigid IT infrastructure soon became apparent.
As the company expanded, so did the number of concurrent Visiopharm users. In parallel, developments in Artificial Intelligence (AI) powered image analysis technologies translated into an increased demand for GPU-driven Deep Learning algorithms. In response to this, we migrated our IT infrastructure to a purpose-built AWS cloud environment, with flexibility, scalability and speed at the heart of it. Our proprietary ‘QDPConnect’ cloud management system, built with our sister company Sciento, enables our Image Analysis scientists to launch virtual machines containing Visiopharm, with the desired hardware specifications such as GPU support, processing power and memory, to suit specific study requirements.
It’s always a great experience to interact with the Visiopharm team! Their technical experts are always on hand to answer any questions, from supporting installation of the latest software update, to offering advice on specific software issues. A number of our team have also benefited from attending Visiopharm Academy courses to further develop their knowledge of the software. More recently, we joined Visiopharm’s Beta-Tester partner programme, where we get access to the latest software releases and provide expert feedback on the new implemented features and tools, such as the Phenoplex module and the Guided QC workflow.
It’s always a great experience to interact with the Visiopharm team! Their technical experts are always on hand to answer any questions, from supporting installation of the latest software update, to offering advice on specific software issues.

Visiopharm: What kind of problems or questions of your customers can you address with Visiopharm?
OracleBio: We use Visiopharm on a daily basis to provide answers to the research questions posed by our customers. This can range from a simple tumour-stroma classifier on a single-plex chromogenic IHC study, to more complex cell detection and phenotyping on high-plex immunofluorescence assays.
The Tissuealign™ module within Visiopharm has been very valuable when clients have requested segmentation of the tumour microenvironment of IHC and IF images in the absence of a specific tumour marker or any distinctive tumour morphology. Co-registration of the stained tissue images with their corresponding H&Es has enabled us to establish more accurate segmentation which has subsequently led to the generation of more robust data.
Our clients are often interested in the downstream application of bioinformatics. The flexibility in custom-built apps allows us to generate a large amount of useful information defined by the output variables; from a simple cell count (including positive and negative cell populations) to specific x-y coordinates for each cell and mean intensity data for all marker layers present. The exported data can be compiled into formats compatible with a client’s bioinformatics platform for subsequent data analysis.
Visiopharm: What specific features of our product do you find most helpful?
OracleBio: The flexibility in app development is one of the most useful features of Visiopharm and allows us to undertake a variety of projects ranging from single-plex IHC to challenging multiplex IF analysis. Being able to custom-build algorithms to address image analysis challenges is a major advantage of the software. There are a number of routes to take during algorithm development and depending on the tissue type, marker staining and required outputs, algorithms can be easily adapted to optimise the results for each study.
The flexibility in app development is one of the most useful features of Visiopharm and allows us to undertake a variety of projects ranging from single-plex IHC to challenging multiplex IF analysis.
More specifically, the addition of AI to the software has greatly improved our experience, especially with regards to nuclei segmentation. Variable stain intensity is a common factor seen within and between batches of samples, predominantly in clinical studies. The implementation of AI tools has allowed us to develop more robust algorithms that can be applied across a greater range of staining and tissue types. Additionally, the continual software updates, which have recently included improvement to marker visualization and the usability of the colour adjustment tools, provide an enhanced overall user experience.
Visiopharm: Can you share any recent results/outcomes that you have achieved using the platform?
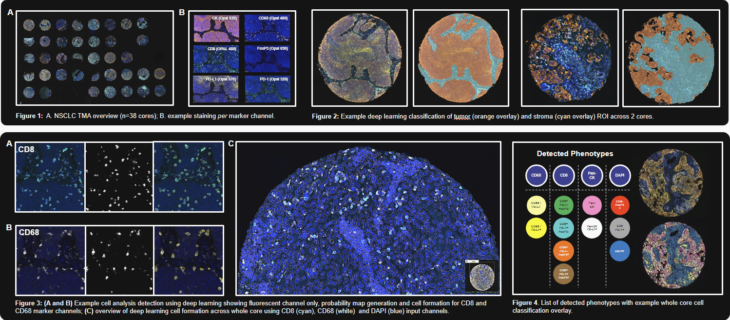
OracleBio: Last year, our collaborative poster with Akoya Biosciences was presented at AACR 2023. OracleBio performed image analysis on samples stained with one of Akoya’s signature mIF panels. We used Visiopharm to profile the immune contexture and tumour spatial interactions in a set of tissue cores, covering a range of NSCLC subtypes and tumour staging. Our approach started with the generation of CD8, CD68 and DAPI cell objects using a Deep Learning algorithm trained on a range of TMA cores. A hierarchical post processing approach was then applied to create CD8, CD68, PanCK and remaining DAPI cell objects for downstream phenotyping. We generated cell object data per core using Visiopharm and carried out spatial analysis using an OracleBio proprietary program (PhenoXplore) to perform neighbourhood analysis for selected phenotypes.
For more details on how we’ve used Visiopharm to analyse samples by incorporating a hierarchal approach to cell detection and phenotyping, check out this case study where we analysed 8-plex multiplex IF staining in CRC & NSCLC tissue. Tissue segmentation using AI deep learning is also demonstrated within Pathology-defined tumour microenvironment regions.
Another poster, presented at the Digital Pathology & AI Congress US 2022, demonstrates how we utilised Visiopharm to develop image analysis techniques to quantify HBV on liver samples. The study samples were stained using a 12-plex multiplex IF assay to provide a better understanding of the liver viral burden and immune cell types involved in this disease. To ensure the most accurate detection of all required phenotypes, customized cellular analysis algorithms were developed to detect and phenotype individual cells. T-cells (CD3) and B-cells (CD20) were identified, along with macrophages (CD68) and these populations were further classified to identify immune cell phenotypes of interest. Hepatocytes were formed using the NaK-ATPase membrane marker and grouped into HBcAg-positive and HBsAg-positive populations. The workflow demonstrates the capabilities of Visiopharm in tackling high-plex studies where precise segmentation of sub-cellular structures is vital to generate robust data and support our clients’ needs.
Since 2020, OracleBio has been a partner in the INCISE project, a triple helix collaboration between industry, academia and the UK’s National Health Service. The goal of the project is to develop an AI-powered risk stratification tool that will predict polyp recurrence using data from pathology, genomics, transcriptomics and clinical records. As the Digital Pathology lead, OracleBio was responsible for configuring the image analysis pipeline, which was built around Visiopharm. Using the software’s Deep Learning capabilities, we build, train and run a selection of apps, including tissue classifiers and cell detection algorithms, on IHC and multiplex sections. Thanks to Visiopharm’s built-in parallel processing feature, supported by our scalable AWS cloud infrastructure, we are able to concurrently process multiple apps across several sections from the 2,700-patient cohort at speed, and deliver the results back to the consortium in a timely manner.

Transform Your Research with Cutting-Edge Image Analysis
As we wrap up the overview of OracleBio’s journey with Visiopharm, it’s clear that this collaboration brings pivotal benefits to the CRO and its clients. Key advantages include:
Robust Data Generation: The integration of Visiopharm’s software into OracleBio’s processes has enabled the generation of more accurate and reliable data from histology images, enhancing the quality of R&D outcomes for their clients.
Tailored Analytical Capabilities: Visiopharm’s flexible software allows OracleBio to develop customized image analysis algorithms. This adaptability is crucial for meeting the diverse and specific needs of their clients in various research areas.
Expanded Operational Capacity: The adoption of Visiopharm’s technology, particularly its cloud-compatibility, has significantly increased OracleBio’s analytical capacity. This allows for handling larger-scale projects more efficiently, thereby expanding their service offerings and client base.
Are you curious to hear more about our image analysis software? Don’t hesitate to contact us for more information.
For more information about OracleBio’s services, contact the team at enquiries@oraclebio.com



