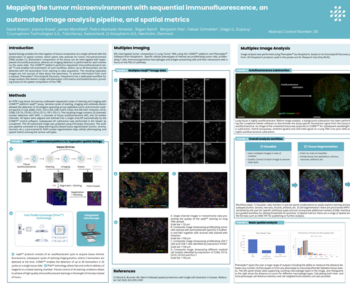
A novel global phenotyping method using artificial intelligence to overcome the cell segmentation bottleneck


Multiplex methods for the detection of protein expression generate extremely data-rich images of intact tissue sections. These images are invaluable for the quantification and analysis of complex biology and have huge potential for biomarker development. However, their interpretation presents a considerable data analysis challenge which limits their utility, especially in clinical workflows.
The key task from these images is often cellular phenotyping – e.g. the identification of immunophenotypes, or the separation cells with compartment-specific protein expression from others. Most current automated methods depend upon a cellular segmentation step to support this, and segmentation has become an area of intensive research in AI (Artificial Intelligence), but the task is very difficult and no universally accepted method has been identified.
Therefore, methods of cellular phenotyping which operate without the need for segmentation would be very valuable. To meet this challenge, we have developed an AI cellular phenotyping method which does not require segmentation at all.
We use the U-Net backbone within the Visiopharm deep learning module to train our algorithm using expert human annotations of entire cellular regions. Crucially, we need only train for a single example of each compartmental stain (nuclear/cytoplasmic/membranous), and the algorithm then assigns class identities to cells without the need for segmentation of nuclei cytoplasm or membranes, using regional information in a manner analogous to a human expert, who may not need to be confident about cellular boundaries to accurately phenotype a cell.
The method is comparable in accuracy to segmentation-based methods, is highly transferrable, and represents a highly novel approach to quantitative image analysis with the potential to radically alter the way that we analyze these data-rich images.

Rachel Baird (née Pennie), MSc
Rachel Baird, MSc is a translational histologist in the deep phenotyping advanced technology facility within the Le Quesne Lab at the CRUK Scotland Institute. She is an expert in the development and application of Visiopharm deep learning methods to multiplex and high plex histopathological images.