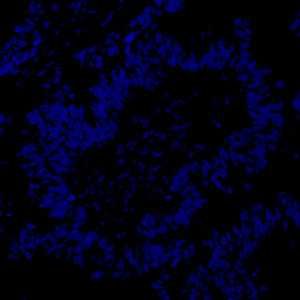
Tissue stained with DAPI
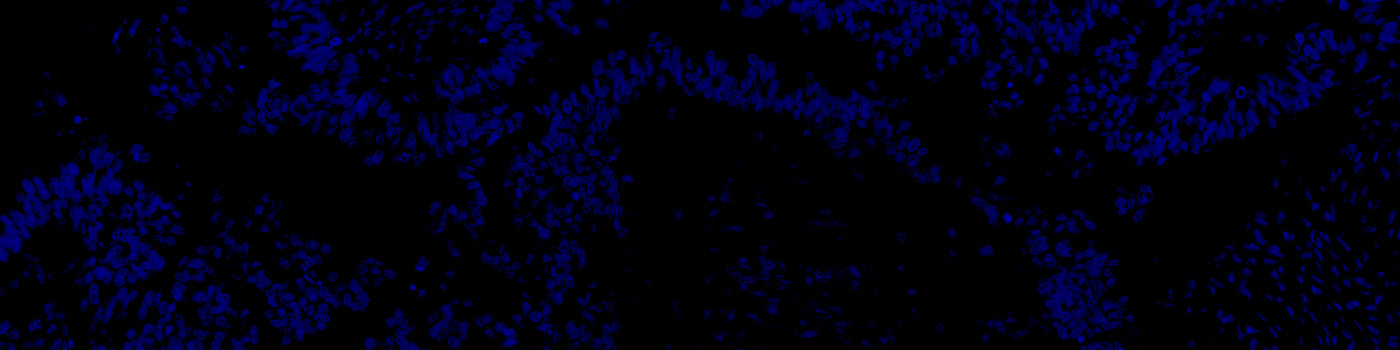
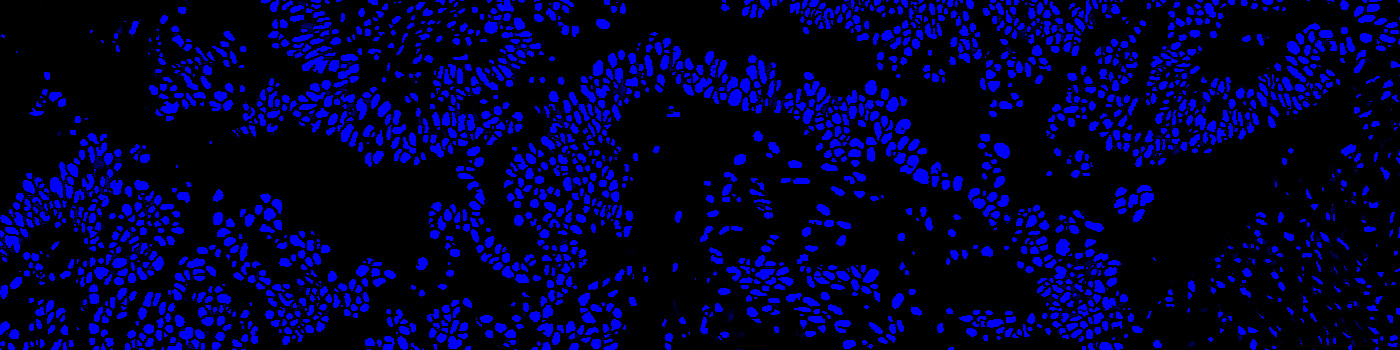
#10169
Developed for automatic segmentation of nuclei in fluorescence
Identification and segmentation of individual nuclei is of interest in many applications. Nuclei can be difficult to detect accurately and precisely across different images using traditional image analysis with feature engineering. This APP utilizes artificial intelligence (AI) for automatic nuclear segmentation in fluorescence images stained with a family of blue dyes for DNA identification. It consists of a pre-trained deep neural network and the APP is ready for use without additional training.
The APP functions as a template where the user can easily configure it to fulfill their needs using the Visiopharm AuthorTM module. The APP currently has one configuration option which can be switched on and off as needed:
It is also possible for the user to customize the APP by adding other post-processing steps and/or output variables as needed.
Quantitative Output variables
The output variable obtained from this protocol is:
Workflow
Step 1: Load an image
Step 2: Load and run the APP “10169 – Nuclei Detection, AI (Fluorescence)”
Methods
The APP was trained using 41,000 annotated nuclei from DAPI stained tissue. The architectural structure of the network is a U-Net which is popular for medical image segmentation. The neural network uses a cascade of layers of nonlinear processing units for feature extraction and transformation, with each successive layer using the output from the previous layers as input. U-Net uses an encoder-decoder structure with a contracting path and an expansive path. For more information of the network architecture, see [1].
Staining Protocol
Tissue must be labeled with a blue fluorescent dye for DNA staining such as DAPI, Hoechst or Iridium such that all nuclei are labeled.
Additional information
To run the APP, a NVIDIA GPU with minimum 4 GB RAM is required.
The APP utilizes the Visiopharm Engine™, Engine™ AI and Viewer software modules, where Engine™ and Engine™ AI offer an execution platform to expand processing capability and speed of image analysis. The Viewer allows a fast review together with several types of image adjustment properties e.g. outlining of regions, annotations and direct measures of distance, curve length, radius, etc.
By adding the Author™ and/or Author™ AI modules the APP can be customized to fit other purposes. These modules offer a comprehensive and dedicated set of tools for creating new fit-for-purpose analysis APPs, and no programming experience is required.
Keywords
DAPI, Hoechst, Iridium, Fluorescence, Nuclei Detection, Nuclei Segmentation, Deep Learning, Artificial Intelligence, Image Analysis
References
[1] Ronneberger, O. et al. U-Net: Convolutional Networks for Biomedical Image Segmentation. International Conference on Medical Image Computing and Computer-Assisted Intervention, 2015, 234-241, DOI: https://link.springer.com/chapter/10.1007%2F978-3-319-24574-4_28