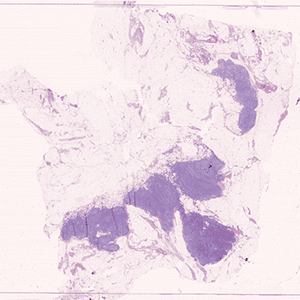
The APP analyses H&E stained whole slide images of lymph nodes associated with breast or colorectal cancer with no need for manual interaction.
#10159
Developed for metastasis detection in H&E stained lymph nodes
Finding metastases in H&E stained lymph node sections can be time consuming and challenging. The differences between small metastases, epithelial tissue and clusters of macrophages are often subtle, and would not be possible to distinguish using conventional image analysis techniques.
This APP utilizes AI/deep learning and has been trained to detect metastases in lymph nodes associated with breast and colon adenocarcinoma, stained with H&E. The deep learning architecture allows it to recognize complex structures and interpret the tissue context when analyzing an image, making it an efficient tool for detecting even small metastases that are not easily noticed.
Quantitative Output variables
The output variables obtained from this protocol are:
Workflow
The APP contains four protocols:
Step 1: Tissue Detect: Outlines tissue on the slide for further analysis.
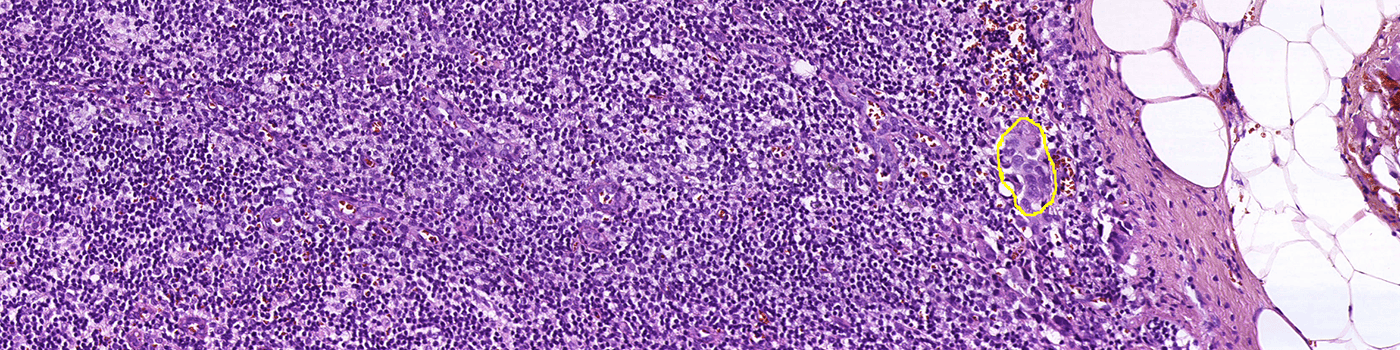
Step 2: Metastasis Detection: Identifies possible metastases using AI.
Step 3: Post Processing: Post-processes the classification results, improving accuracy and visualization.
Step 4: Calculate Results: Locates the single largest metastasis and calculates the diameter and/or calculates the total metastasis area.
The protocols can be run separately or as one using the APP sequence functionality in VIS.