Image of H&E stained needle biopsies from prostate tissue to be analyzed.

#10160
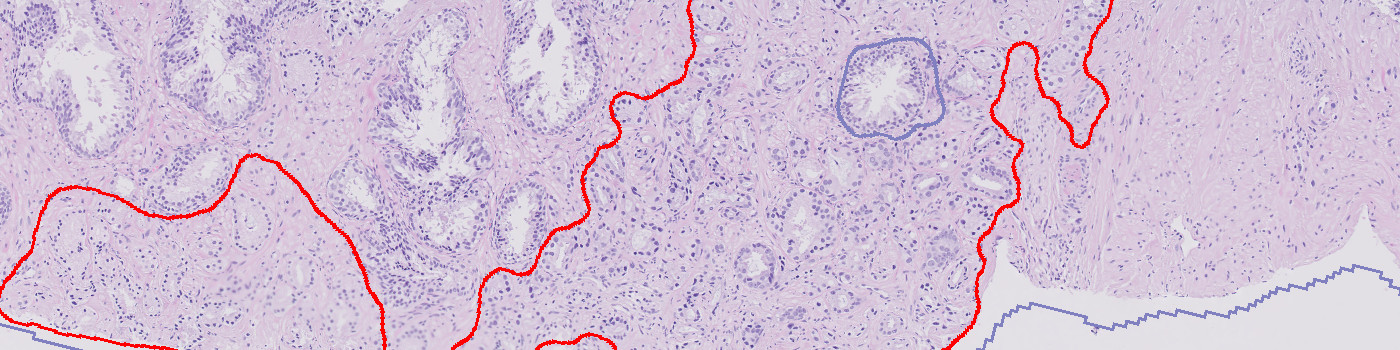
Developed for tumor detection in H&E stained prostate tissue
Prostate cancer is the second most common cancer in men, with an estimated 1.1 million diagnoses worldwide in 2012, accounting for 15% of all cancers diagnosed [1]. Research in prostate cancer is important to help improve diagnosis and choosing the best treatments for individuals at all stages of the disease. Automated tumor detection can help identify regions-of-interest and provide numerical data in a scalable fashion.
This APP utilizes AI/deep learning and has been trained to detect tumors in images of prostate tissue stained with H&E. The deep learning architecture enables the APP to recognize complex structures and interpret the tissue context when analyzing an image, making it an efficient tool for detecting even small tumors that are not easily noticed. The APP does not grade the tumors.
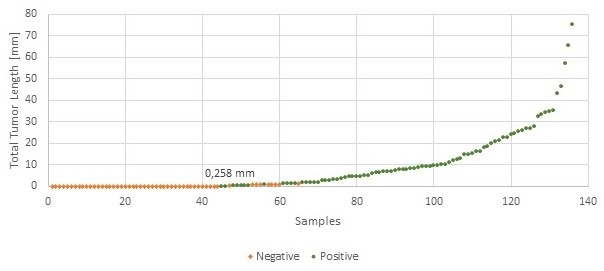
136 slides were manually assessed and classified as positive or negative, resulting in 53 negative and 83 positive slides. The same slides were analyzed with the APP where the Total Tumor Length with a cut-off of 0 mm was used to classify slides as positive and negative. The agreement between manual and APP slide classification is shown below.
| Sensitivity | 97.9 % |
|---|---|
| Specificity | 83.0 % |
Quantitative Output variables
The output variables obtained from this protocol are:
Total Tumor Area [mm²]
Total Tumor Length [mm]
Tumor Percentage [%]
Workflow
The APP contains four protocols:
The principle of procedure for the APP is as follows:
Methods
The APP was developed using the DeepLabv3+ neural network available with Author™ AI. The neural network uses a cascade of layers of nonlinear processing units for feature extraction and transformation, with each successive layer using the output from the previous layer as input. DeepLabv3+ uses an encoder-decoder structure with atrous spatial pyramid pooling (ASPP) that is able to encode multi-scale contextual information by probing the incoming features with filters or pooling operations at multiple rates and multiple effective field-of-views.
This means that instead of using stepwise upsampling blocks to incorporate features from different levels, this network only needs two upsampling steps, i.e. it is faster to train and analyze than e.g. the U-Net. All of this also means that the decoder module can refine the segmentation results along the object boundaries more precisely. For more information on the network architecture, see [2].
Staining Protocol
There is no staining protocol available.
Additional information
To run the APP, a NVIDIA GPU with minimum 4 GB RAM is required.
Keywords
Prostate cancer, H&E, Hematoxylin, Eosin, Deep learning, AI, Image analysis, DeepLabv3+, Adenocarcinoma
References
LITERATURE
USERS
The APP was developed in collaboration with University Medical Center Groningen, The Netherlands.