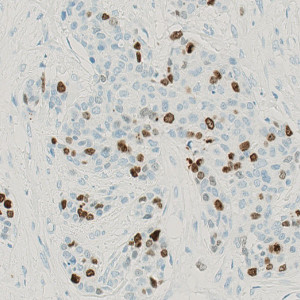
Breast tissue stained with Ki-67 where tumor is present.

#10162
This Quickstart APP will outline tumors and stroma regions with ease across a range of tissues in IHC-stained images. All you need to do is have images to analyze, and the APP will quickly perform a task that once took hours.
Once the APP has completed processing, you will obtain results quantifying the total area of the tumor regions analyzed. The next step in your analysis can be to perform nuclei segmentation with one of the two Quickstart APPs available for this purpose (see the “Related APPs” section below). With batch analysis, both APPs can be queued together on the same image to provide true walk-away analysis.
As with all of our Quickstart APPs and custom-build APPs, your image analysis can be scaled by queueing APPs to run in the background on an unlimited number of images—this is possible with the “batch analysis” feature available on both Discovery and Phenoplex™.
Working with multimodal data sets or TMAs? No problem. Run these APPs after using Tissuealign™ feature to align your images at the cellular level, or the Tissuearray™ feature to automate de-arraying of your tissue cores.
Quantitative Output variables
Tumor Area [mm2]
Workflow
After loading your image, follow these steps:
Step 1: Open the “10162 – IHC, Tumor Detection, AI” APP.
Step 2: Select a region of interest or skip to the next step.
Step 3: Click “Run APP” or queue it with batch analysis and continue working on other images.