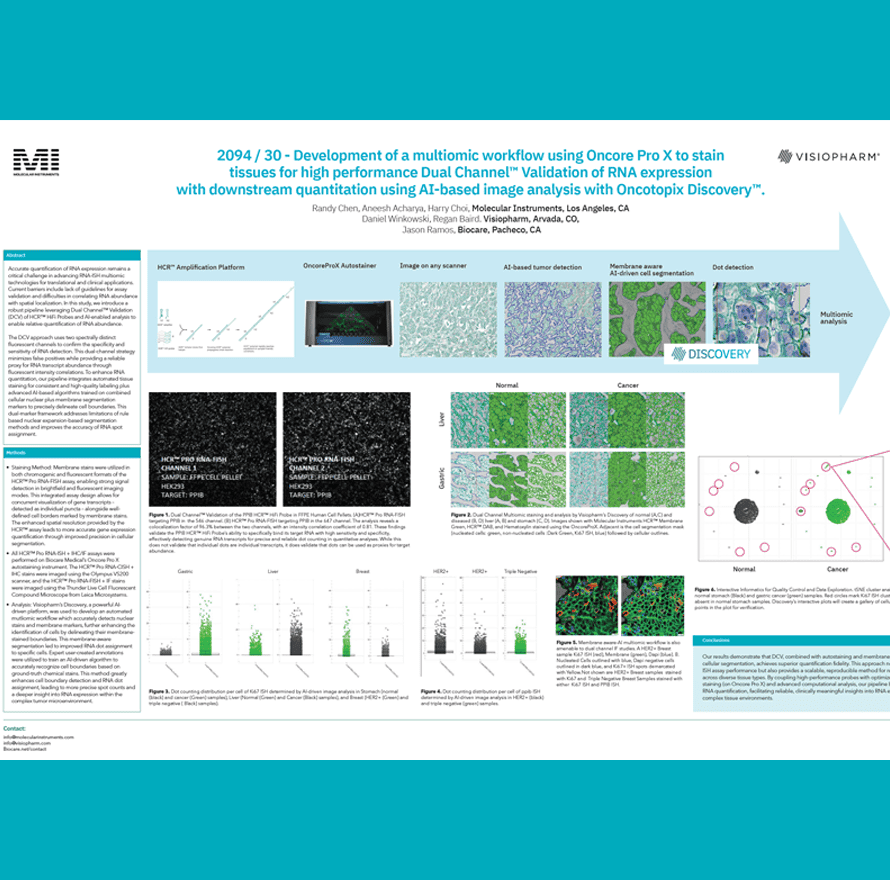

Accurate quantification of RNA expression remains a critical challenge in advancing RNA-ISH multiomic technologies for translational and clinical applications. Current barriers include lack of guidelines for assay validation and difficulties in correlating RNA abundance with spatial localization. In this study, we introduce a robust pipeline leveraging Dual Channel™ Validation (DCV) of HCR™ HiFi Probes and AI-enabled analysis to enable relative quantification of RNA abundance.
The DCV approach uses two spectrally distinct fluorescent channels to confirm the specificity and sensitivity of RNA detection. This dual-channel strategy minimizes false positives while providing a reliable proxy for RNA transcript abundance through fluorescent intensity correlations. To enhance RNA quantitation, our pipeline integrates automated tissue staining for consistent and high-quality labeling plus advanced AI-based algorithms trained on combined cellular nuclear plus membrane segmentation markers to precisely delineate cell boundaries. This dual-marker framework addresses limitations of rule based nuclear expansion-based segmentation methods and improves the accuracy of RNA spot assignment.
Randy Chen1, Aneesh Acharya1, Harry Choi1, Daniel Winkowski2, Regan Baird2, Jason Ramos3
- Molecular Instruments, Los Angeles, CA
- Visiopharm, Visiopharm, Arvada, CO
- Biocare, Pacheco, CA