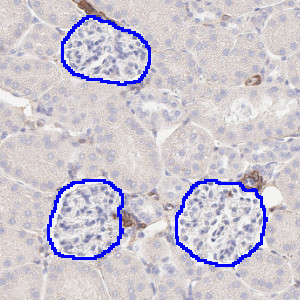
Segmented glomeruli in kidney samples stained with aSMA.
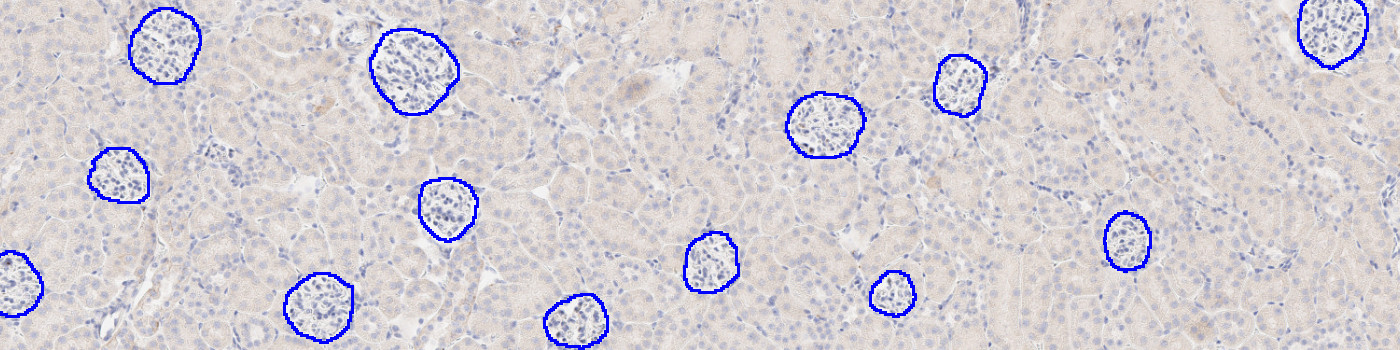
#10145
Developed for automatic segmentation of glomeruli in IHC-stained kidney tissue, this Quickstart APP speeds up the development of custom algorithms for analyzing kidney tissue. This APP automatically identifies and segments glomeruli in kidney samples stained with various biomarkers including CD68, COL1A1, COLIV, synaptopodin, nephrin, WT1 and/or aSMA. It utilizes deep learning for a robust segmentation.
As with all of our Quickstart APPs and custom-build APPs, your image analysis can be scaled by queueing APPs to run in the background on an unlimited number of images and cores—this is possible with the “batch analysis” feature available on both Discovery and Phenoplex™.
Working with multimodal data sets or TMAs? No problem. Run these APPs after using Tissuealign™ feature to align your images at the cellular level, or the Tissuearray™ feature to automate de-arraying your cores.
Quantitative Output variables
Total glomeruli count
Workflow
After loading your image follow these steps:
Step 1: Open the “10145 – IHC, Glomerulus Detection” APP
Step 2: Click “Run APP” or queue this APP with batch analysis and keep working on other images
Additional information
To run the APP, a NVIDIA GPU with minimum 4 GB RAM is required.