Decoding the Liver Microenvironment: A Conversation with Prof. Heather Stevenson-Lerner

The liver’s microenvironment holds the key to understanding and addressing complex liver diseases, from fibrosis to emerging infections. In this blog, we sit down with Professor Heather Stevenson-Lerner, MD, PhD, a physician-scientist and the sole liver pathologist at the University of Texas Medical Branch (UTMB), whose work spans over a decade of groundbreaking research in liver pathology and immune responses.
Prof. Stevenson-Lerner shares her experience leveraging Visiopharm software to explore the liver microenvironment, offering unique insights into macrophage-mediated immune responses and the use of advanced AI tools for spatial biology analysis.
She also highlights how Visiopharm empowers her team to study liver diseases with unprecedented precision, ensuring reproducible and impactful research outcomes.
Read on for the full interview or watch the video recording to discover how Prof. Stevenson-Lerner is driving innovation in liver pathology.
Visiopharm: Welcome, Prof. Heather, can you tell us a little bit about yourself?
Prof. Heather Stevenson-Lerner: My name is Heather Stevenson-Lerner, I’m a professor in the Department of Pathology at the University of Texas Medical Branch (UTMB) in Galveston, Texas, and a physician-scientist studying the liver microenvironment and various types of chronic liver disease. I’ve been using Visiopharm software for nearly seven years, generating valuable data for our projects. I’m excited to share more about our work today.
Visiopharm: Can you describe the primary focus of your research and the specific pathologies you’re studying with Visiopharm software?
Prof. Heather Stevenson-Lerner: I’ve been the sole liver pathologist at UTMB since 2014, signing out every liver biopsy for over a decade. Visiopharm’s tools allow us to study the hepatic microenvironment using formalin-fixed, paraffin-embedded tissue. With access to a 30-year archive of liver tissue, we’ve analyzed a range of chronic liver diseases, such as steatotic liver disease, viral hepatitis C, autoimmune hepatitis, primary biliary cholangitis, and even infectious diseases like Ebola. At UTMB, we specialize in emerging infections, and the ability to analyze deactivated tissues without requiring high-containment labs has been invaluable.
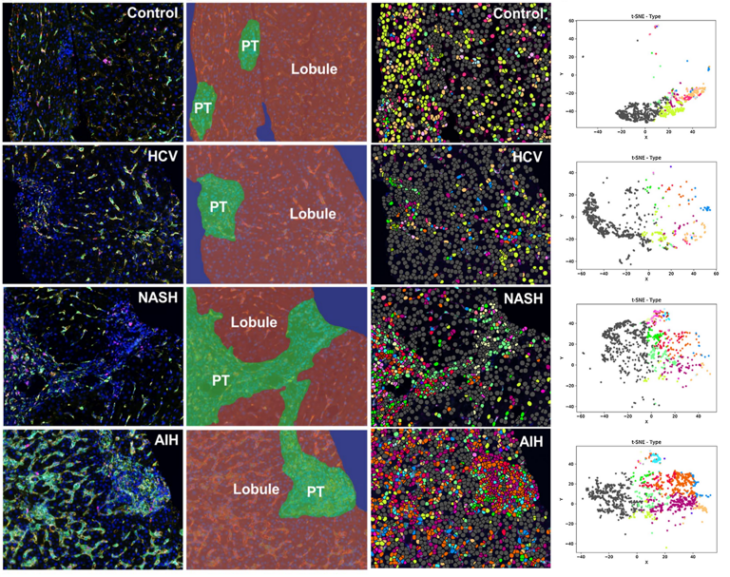
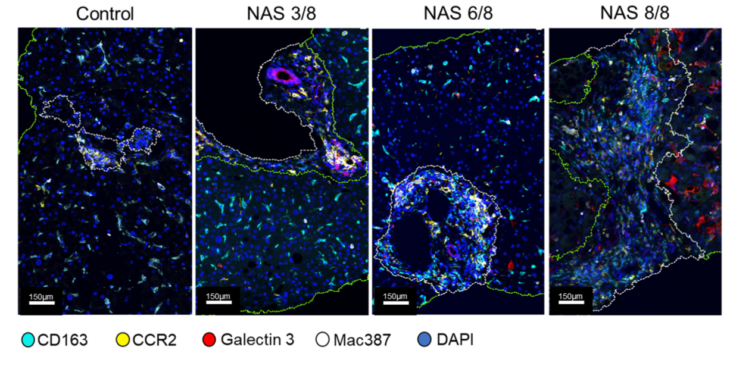
The primary focus of our research is to study the hepatic microenvironment. We’re very interested in macrophage-mediated immune responses, which play a critical role in promoting inflammation and fibrosis in the liver. We use archived liver biopsies to stain different antibodies, and Visiopharm is integral to our work, especially for multiplex staining. For instance, we use a six-antibody panel for macrophages, and we look at markers in the same compartment on the same cell, and there is no other software platform that allows us to analyse that.
“Over the years, we’ve tried a lot of different things, and we found that Visiopharm is the best at helping us differentiate these different phenotypes within the hepatic microenvironment and these various diseases.”
Visiopharm: What challenges or limitations in your research led you to consider using AI solutions like Visiopharm, for example?
Prof. Heather Stevenson-Lerner: I collaborated with Dr. Jared Burks at MD Anderson Cancer Center when we started. Spatial approaches were already well-established for tumor microenvironments, so I thought, why not apply similar techniques to inflammatory liver diseases? We were among the first ones doing it here at UTMB, and now it’s really caught on. Obviously, the spatial field has grown so much in the last few years, but Dr. Jared Burks and I both came to the conclusion that Visiopharm was the best.
“At that time, we tested several different platforms. Each had pros and cons, but Visiopharm stood out, particularly for its AI-driven cell segmentation. Accurate cell segmentation at the initial step is crucial since it ensures precise downstream data analysis.”
In addition, when we first started using Visiopharm, we wanted to make sure that the data was reproducible. You know, patients are different. We have to make sure what we’re seeing is accurate. To validate our results, we collaborated with the bioinformatics group at the University of Michigan, which is one of the best in the country. We compared results from the same experiments using two different analysis methods: multicomponent TIFF files processed with Visiopharm and numerical cell segmentation data from InForm software (Vectra Polaris). And we did come up with the same. We have the same results by looking at the data in two completely different ways. The advantage of Visiopharm is that we can perform the analysis independently in our own lab without relying on external support.
Visiopharm: How has the use of Visiopharm software improved the quality and accuracy of your research outcomes?
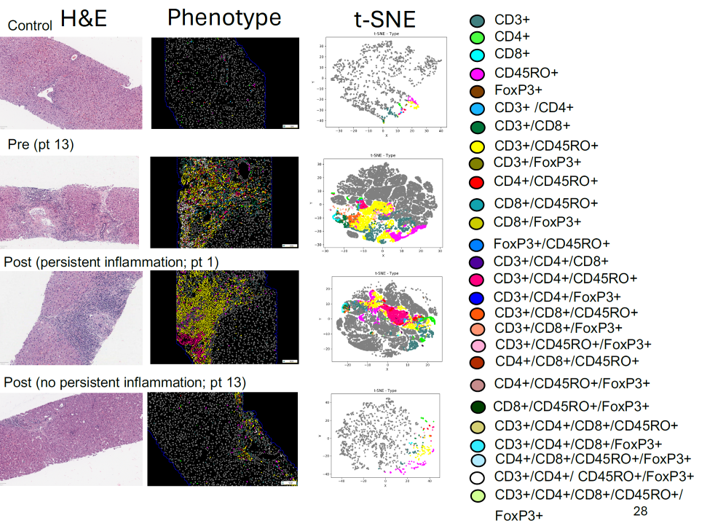
Prof. Heather Stevenson-Lerner: We’re really trying to preserve the spatial aspects and keep the liver architecture intact. There are many great things out there. For example, when I was a graduate student, I did flow cytometry in the liver. But with flow cytometry, you have to homogenize the entire liver tissue. First, you need fresh tissue—not formalin-fixed—which can be challenging, especially with human patients. When you homogenize the liver, you separate the cells but can’t determine their spatial location. For example, is a cell in the portal tract or the lobule of the liver? What types of cells are next to each other? Who is interacting with whom? You can’t perform this kind of neighbor analysis with flow cytometry.
Similarly, single-cell RNA sequencing recreates the liver microenvironment but still doesn’t preserve the architecture. Visiopharm allows you to analyze different cell types while maintaining spatial relationships. You can identify phenotypes, measure marker expression levels, and perform tSNE to study the relationships between cells. This ability to preserve liver architecture is something that the Visiopharm analysis enables that other applications cannot. Additionally, when looking for small signals, we also use NanoString to analyze whole RNA signatures from homogenized liver tissue. While NanoString is powerful, homogenization can dilute specific signals, such as those from a single cell type with a druggable target or marker expression. With Visiopharm, I can go directly into the liver tissue, segment CD68-positive macrophages, cluster them, and analyze their signals. This allows me to identify data I might miss when homogenizing the entire liver, making Visiopharm an incredibly powerful tool.
Visiopharm: In which ways did Visiopharm software prove to be the only viable solution for your research needs compared to other tools or methods you considered?
Prof. Heather Stevenson-Lerner: Well, I think there are several important things. As I mentioned, being able to stain multiple antibodies in the same cellular compartment is crucial. We try to combine different types of antibodies—some that are nuclear, cytoplasmic, or membranous—which helps. However, we also work with antibodies like CD68, CD163, CD14, and CD16, which are all in the same compartment but labeled with different fluorophores or Opal fluorophores. Visiopharm has been the best at accurately separating these cells with antibodies in the same compartment.
Another key advantage is the ability to use pre-existing algorithms from Visiopharm’s website. Once we develop customized algorithms, we can apply them to different chronic liver diseases with minimal adjustments. For example, we created an algorithm for chronic hepatitis C (HCV) and, with slight modifications, applied it to patients with autoimmune hepatitis. This flexibility is especially helpful when using the same macrophage antibody panel across various diseases.
Additionally, the training sessions with Visiopharm have been invaluable. These sessions are included in our software package, and we’ve had weekly meetings during heavy usage periods or when addressing software updates or specific questions. This ongoing training has been crucial to our success.
“Finally, Visiopharm’s customer support has been excellent. They are always available to assist whenever we have questions, ensuring we’re never held up in our research. These aspects—antibody differentiation, customizable algorithms, training, and support—are the main things that have been extremely helpful for us.”
Are you curious to see the work of Prof. Heather Stevenson-Lerner? Check out these recent publications:
Saldarriaga OA, Wanninger TG, Arroyave E, Gosnell J, Krishnan S, Oneka M, Bao D, Millian DE, Kueht ML, Moghe A, Jiao J, Sanchez JI, Spratt H, Beretta L, Rao A, Burks JK, Stevenson HL. Heterogeneity in intrahepatic macrophage populations and druggable target expression in patients with steatotic liver disease-related fibrosis. JHEP Rep. 2023 Nov 3;6(1):100958. doi: 10.1016/j.jhepr.2023.100958. PMID: 38162144; PMCID: PMC10757256.
Wanninger TG, Saldarriaga OA, Arroyave E, Millian DE, Comer JE, Paessler S and Stevenson HL (2024) Hepatic and pulmonary macrophage activity in a mucosal challenge model of Ebola virus disease. Front. Immunol. 15:1439971. doi: 10.3389/fimmu.2024.1439971


