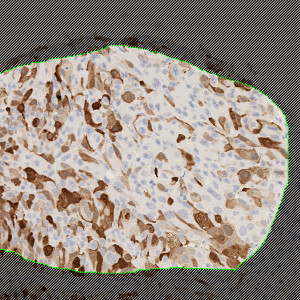
Image of a manually outlined tumor region in a bladder tissue TMA core stained by IHC for UBE2C.
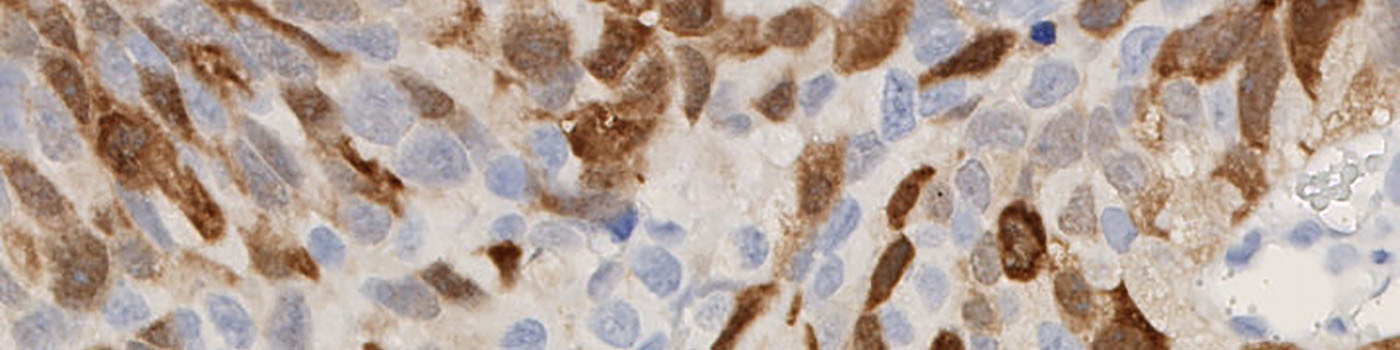
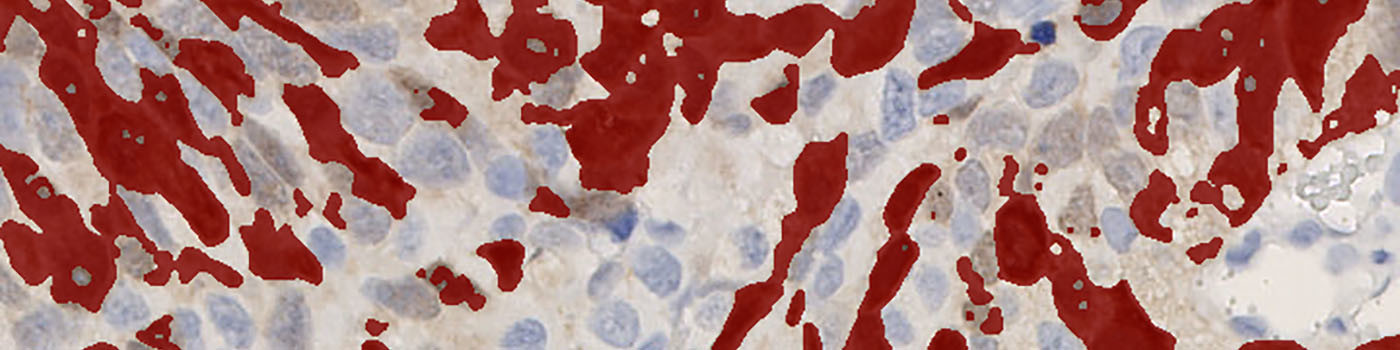
#10016
UBE2C is only expressed at low levels in normal tissues, see [1]. However, the expression of UBE2C is high in cancerous tissues of various origins, see [2]. High level of UBE2C expression is prognostic of poor survival in colorectal, thyroid, breast, see [3], ovarian cancer, see [4], lymphoid neoplasm, and in non-small cell lung cancer, see [5]. UBE2C might be a potential marker for tumor response to therapy when down-regulation is detected and the potential of UBE2C as a therapeutic target in cancer should be further explored, see [2].
This APP can be used for quantifying the cytoplasmic expression of UBE2C. A statistically significant correlation to manual scoring, in a study involving 300 patients was found [6].
This APP was developed for, and validated by, Niels Fristrup MD and Dr. Lars Dyrskjøt Andersen, Center for Molecular Clinical Cancer Research Department of Molecular Medicine (MOMA) Aarhus University Hospital.
Quantitative Output variables
The output variables obtained from this protocol include:
Methods
The method used for computing the UBE2C expression is based on a smoothed color-contrast between the red and the blue color band. An intensity threshold is used to determine positivity and negativity of the cytoplasmic expression.
From the segmentation, the average DAB intensity across all positive cytoplasm areas is measured and subtracted from 255. This is done in to associate high values with a high staining intensity, which is more intuitive.
It should be noted that this approach to quantifying biomarker expression is vulnerable to variations in protocols for tissue preparation (fixation time, tissue thickness, reagents, auto-stainer type and settings, etc.). Furthermore, the application of this APP for TMA study designs is robust within the TMA.
Keywords
UBE2C, bladder cancer, metastasis, immunohistochemistry, image analysis, quantitative, digital pathology.
References
LITERATURE
1. Okamoto, Y., et. al. UbcH10 is the cancer-related E2 ubiquitinconjugating enzyme, Cancer Res. 2003, 63 (14), 4167-73, DOI
2. Hao, Z., et. al. Ubiquitin-conjugating enzyme UBE2C: molecular biology, role in tumorigenesis, and potential as a biomarker, Tumour Biol. 2012, 33 (3), 723-30, DOI
3. Loussouarn, D., et. al. Validation of UBE2C protein as a prognostic marker in node-positive breast cancer, Br J Cancer 2009, 101 (1), 166-73, DOI
4. Berlingieri, M.T., et. al. UbcH10 expression may be a useful tool in the prognosis of ovarian carcinomas, Oncogene 2007, 26 (14), 2136-40, DOI
5. Kadara, H., et. al. Identification of gene signatures and molecular markers for human lung cancer prognosis using an in vitro lung carcinogenesis system, Cancer Prev Res 2009, 2 (8), 702-11, DOI
6. N. Fristrup, et al. Multicenter validation of cyclin D1, MCM7, TRIM29, and UBE2C as prognostic protein markers in non-muscle-invasive bladder cancer. Am. J. Pathol. 2013, 182 (2), 339-349, DOI