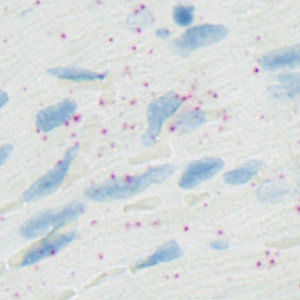
Field-of-view from the left ventricle of a rat heart stained for EGLN3.
#10127
Presented in 2012, the RNAscope is one of the more recent RNA in situ hybridization techniques that allows visualization of multiple target genes. RNAscope uses a probe design strategy that amplifies the signal and suppresses the background, which results in increased sensitivity and specificity, see [1].
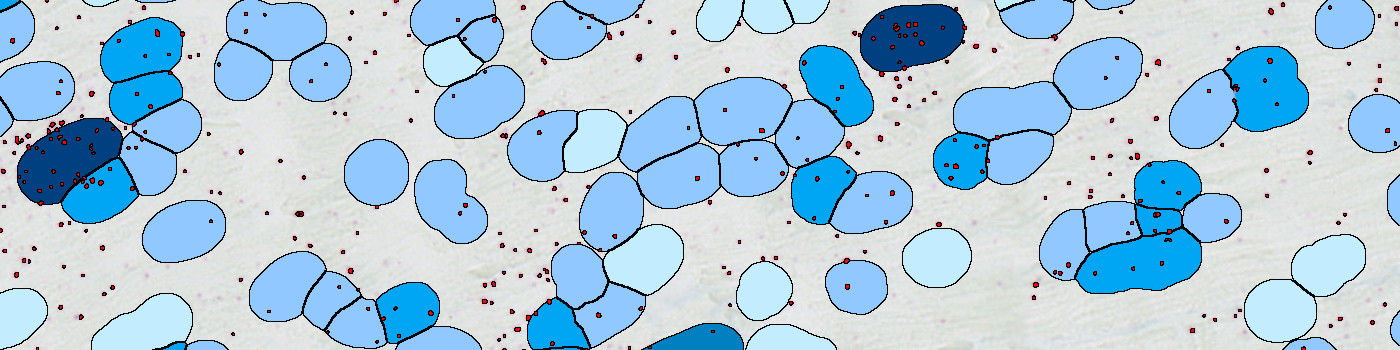
The protocol automatically detects and quantifies probes for a wide range of genes: ANGPT2, EGLN3, GLP1R, HIF1A, PECAM1, SLC2A1 and dapB. Each cell is classified as 0, 1+, 2+, 3+, or 4+ based on the number of probes it contains as recommended by ACD.
Quantitative Output variables
The output variables obtained from this protocol include:
Workflow
Step 1: Manually outline ROIs.
Step 2: Load and run the APP for quantification of probes and cells.
Methods
The protocol works within a manually outlined region of interest (ROI). Initially, probes and nuclei are detected. Probes are identified using a polynomial blob filter and the haematoxylin color-deconvolution band is used for nuclei identification. The initial classification of nuclei and probes is managed with a postprocessing protocol to refine the detection of nuclei and probes and to simulate cytoplasm. Cytoplasm is simulated by extending nuclei a predefined diameter, or until another cell is encountered. Cells are classified as 0, 1+, 2+, 3+, or 4+ based on the number of probes covered.
Staining Protocol
There is no staining protocol available.
Keywords
RNAscope, heart, rat, composite score, digital pathology, image analysis, RNA, ISH, probe, ANGPT2, EGLN3, GLP1R, HIF1A, PECAM1, SLC2A1, dapB.
References
USERS
The APP was developed for Charles Pyke, Camilla Ingvorsen, and Jonas Ahnfelt-Rønne, Novo Nordisk A/S.
LITERATURE
1. Wang, F. et. al. RNAscope – A Novel in Situ RNA Analysis Platform for Formalin-Fixed Paraffin-Embedded Tissues, Journal of Molecular Diagnostics 2012, 14 (1), 22-29, DOI.