
Original TMA-core stained with PCK.
#10017
This APP has been developed to quantify the mitotic index of PHH3 positive cells within breast carcinoma. The APP uses the Virtual Double Staining (VDS) technique, which enables automated and robust detection of tumor regions.
Two serial sections stained respectively for PHH3 and Pancytokeratin (PCK) must be used in this APP. Tumor regions are identified automatically on the PCK stained slide and the outlined tumor region is overlaid on the PHH3 stained slide, thus automatically identifying tumor regions.
Phosphohistone H3 (PHH3) is a mitotic marker and has prognostic capabilities when applied to breast cancer tissue, see [3]. The mitotic index can be obtained by PHH3-immunohistochemical staining, which can be used as a supplement for diagnosis.
PCK is used as a cytoplasmatic marker to localize the cancer tissue.
This APP can be used to automatically outline tumor regions and determine the mitotic index. The APP described here is developed for TMA cores, but the method can easily be applied to whole slides as well.
Auxiliary APPs
Auxiliary APPs are used for additional process steps, e.g. finding Region of Interest (ROI).
Several auxiliary APPs are used for the final nuclei segmentation. These include: Tissue segmentation, Tissue alignment and ROI detection.
Quantitative Output variables
The output variables obtained from this protocol include the number of positive nuclei profiles, the area of tumor tissue and the mitotic index. The mitotic index is calculated as follows:
Methods
To employ the VDS approach for PHH3, two serial sections must be stained for PHH3 and PCK respectively.
This APP works as a three-step process as described below.
First, alignment of TMA cores from the two serial sections is performed (see FIGURE 3). The alignment is done both on a large, whole core scale, and on a finer detail level, to get the best possible match of the two tissue sections.
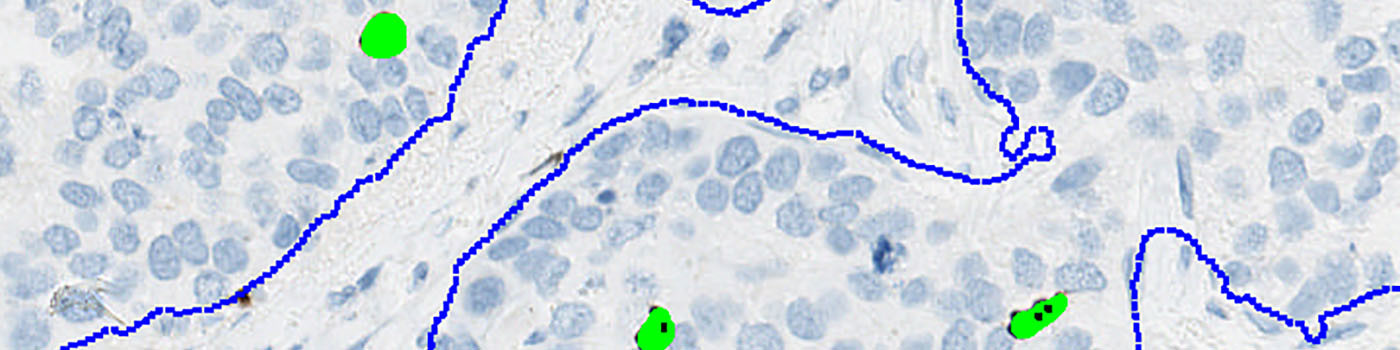
Second, tumor areas are automatically detected from the PCK slide and outlined as regions of interest (ROIs) (see FIGURE 4). The ROIs are then superimposed on the aligned PHH3 tissue slide to outline the tumor region for subsequent analysis limited to the inside of the tumor regions (see FIGURE 5).
Finally, segmentation is performed of all nuclei on the PHH3 stained image inside the ROIs (see FIGURE 6). A method for nuclei separation which is based on shape, size and nuclei probability is used, employing a fully automated watershed-based nuclei segmentation technique. A post-processing step involving a morphological closing operation is applied to avoid counting nuclei in the late mitotic phase (anaphase and telophase) as two nuclei.
The number of positive PHH3 nuclei and the mitotic index are calculated.
Additional information
This APP has been developed in cooperation with Professor Mogens Vyberg from NordiQC and Aalborg University Hospital Denmark and assistant professor Mette Christa Zeuthen from the Danish University College Metropol.
Keywords
VDS, Virtual Double Staining, breast carcinoma, PHH3, Phosphohistone, immunohistochemistry, breast cancer, quantitative, digital pathology, image analysis, PCK, Cytokeratin, mitotic index, TMA
References
USERS
This APP was developed for, and validated by, Niels Fristrup MD and Dr. Lars Dyrskjøt Andersen, Center for Molecular Clinical Cancer Research Department of Molecular Medicine (MOMA) Aarhus University Hospital.
LITERATURE
1. Skaland, I., et. al. Phosphohistone H3 expression has much stronger prognostic value than classical prognosticators in invasive lymph node-negative breast cancer patients less than 55 years of age, Mod Pathol 2007, 20 (12), 1307-15, DOI
2. Kårsnäs, A., et. al. Learning histopathological patterns, Journal of Pathology Informatics 2011, 2 (2), S12, DOI
3. Jung, C., et. al. Segmenting Clustered Nuclei Using H-minima Transform-Based Marker Extraction and Contour Parameterization, IEEE Transactions on Biomedical Engineering 2010, 57 (10), 2600-4, DOI