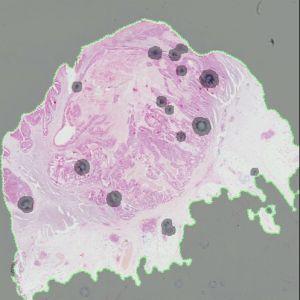
Automated detection of ROI. The area surrounded by the green line is the ROI. Areas that are grayed out are not part of the ROI.
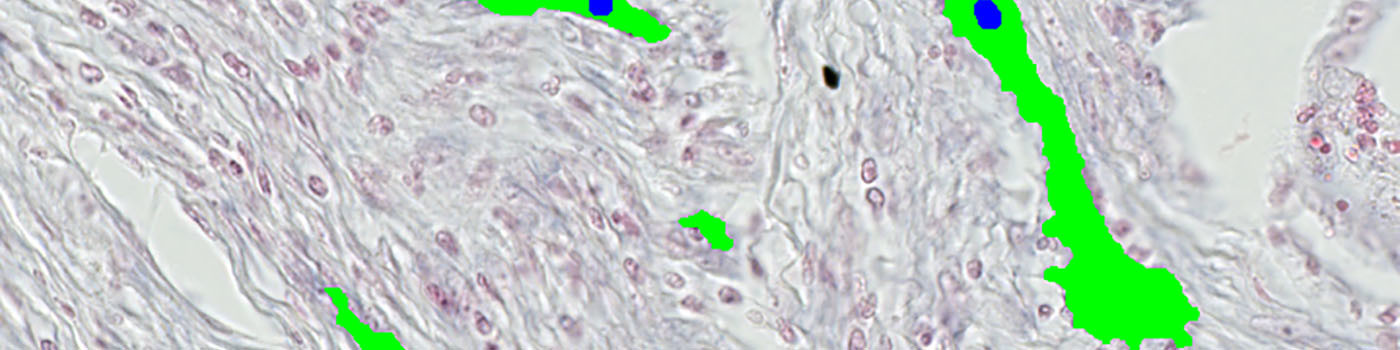
#10024
Angiogenesis is the formation of new blood vessels originating from the endothelium of existing vasculature. Angiogenesis in cancers takes place in parallel with tumor progression. Vascularization facilitates tumor progression and probably also metastasis, and the degree of vascularization may predict drug response by some drug regimens. MicroRNAs constitute a group of non-coding RNAs, which can be detected in tissues by in situ hybridization (ISH). The in situ hybridization assay is based on the use of LNA:DNA chimeric probes that are visualized after chromogenic deposition of the dark-blue precipitate 4-Nitro-blue tetrazolium/5-brom-4-chloro-3-indolylphosphate (NBT/BCIP). A probe for miR-126 is used to detect vessels in colorectal cancers, and sections are counterstained with nuclear fast red to allow orientation in the tissue area and to identify specific tumor areas as regions of interest.
This APP can be used to identify vessels and calculate the microvessel density, cross-sectional area fraction and volumetric intensity measure.
Auxiliary APPs
Auxiliary APPs are used for additional process steps, e.g. finding Region of Interest (ROI).
ROI
The auxiliary APP, ’01 ROI Detect’ can be used to automatically detect the region of interest (ROI). The APP utilizes a Bayesian classifier to detect the tissue areas of interest (see FIGURE 1). In some cases, the operator may subsequently have to manually remove certain irrelevant tissue areas to be excluded from the microvessel quantification, e.g. normal and/or necrotic tissue regions.
Quantitative Output variables
The output variables obtained from this protocol include:
Methods
For an optimal outcome of this APP, it is recommended to start by using the auxiliary APP to detect the ROI (see Auxiliary APPs). This is used for outlining the region of interest (ROI) by identifying the tissue area and discarding staining artifacts (see FIGURE 1).
After outlining the ROI, the main protocol is used for vessel identification (see FIGURE 3). This protocol utilizes the contrast between the red and blue color bands as input and detects the vessels by a threshold classification in the ROIs. Analysis of full virtual slides takes place in a tile-by-tile fashion. If not handled appropriately, vessel profiles that are intersecting with neighboring tile boundaries would be counted twice (or more). Using an unbiased counting frame, see [3], this can be avoided (see FIGURE 4). This principle is implemented in the present APP. Depending on vessel size and density, the application of this principle could make an important difference.
References
USERS
This protocol was configured for: Boye Schnack Nielsen, Bioneer A/S.
LITERATURE
1. Nielsen, B.S., et. al. High levels of microRNA-21 in the stroma of colorectal cancers predict short disease-free survival in stage II colon cancer patients, Clin Exp Metastasis 2011, 28 (1), 27-38, DOI
2. Rask, L., et. al. High expression of miR-21 in tumor stroma correlates with increased cancer cell proliferation in human breast cancer, Apmis 2011, 119 (10), 663-73, DOI
3. Howard, C.V., Reed, M.G. (2005). Unbiased Stereology: Three-dimensional measurement in microscopy. QTP Publications.