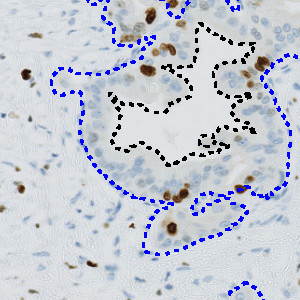
Automatic outline of tumor regions.
#10140
The Ki-67 protein is associated with cellular proliferation, and the protein is present in the nucleus of all cells that are in the active phase of the cell cycle, but absent in resting cells, see [1]. The cell proliferation rate can be assessed by Ki-67-immunohistochemical (IHC) staining, and this can be correlated to the tumor grade and the clinical course. Ki-67 proliferative index is suggested to be an important prognostic variable and is used as a grading parameter for neuroendocrine neoplasms, see [2], including pancreatic neoplasms.
This APP can be used to assess tumors by determining the Ki-67 positivity. The protocol detects and classifies nuclei as positive or negative and returns an average proliferation index for the entire tumor region. Tumor regions must be identified and outlined manually within a region of interest (ROI).
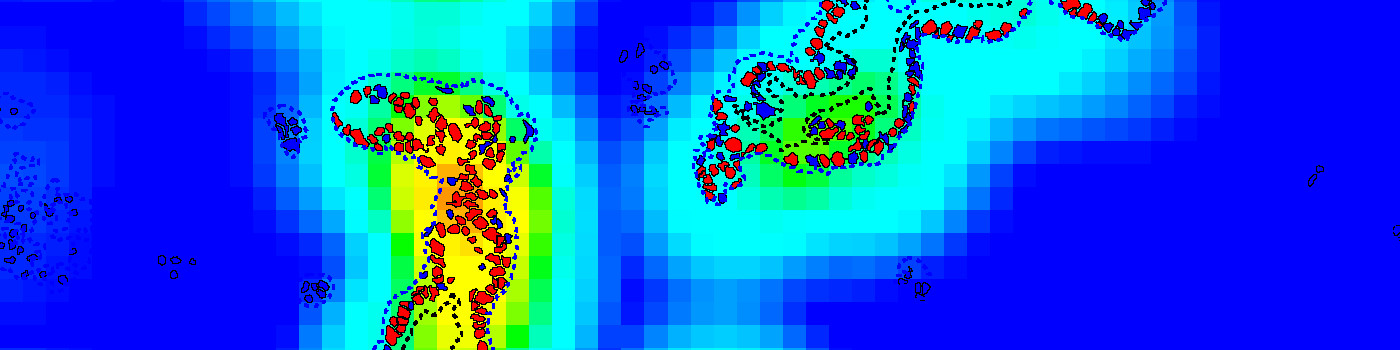
The APP can also be used in a more advanced workflow, including automatic tumor detection and hotspot analysis (Figure 1-5). In this setup, tumor regions are identified automatically from cytokeratin stained slides (e.g. PCK) using the APP “20002 – PCK VDS, Tumor Detection”. The APP “10114 – Hot Spot” is used to generate a heatmap to identify a hot spot containing, e.g. 100 cells and the proliferative index is reported for both the entire tumor region and the hot spot.
Quantitative Output variables
The output variables obtained from this protocol are:
Workflow
SIMPLE
Step 1: Manually outline tumor areas as regions of interest (ROIs)
Step 2: Load and run the APP 10140 – Ki-67, Pancreatic Cancer to analyze the nuclei in tumor region(s)
WITH AUTOMATIC TUMOR DETECTION AND HOT SPOT ANALYSIS
Step 1: Align serial slides of Ki-67 and PCK
Step 2: Load and run the APP 20002 – PCK VDS, Tumor Detection to outline tumor areas
Step 3: Load and run the APP 10140 – Ki-67, Pancreatic Cancer to analyze the nuclei in tumor region(s)
Step 4: Load and run the APP 10114 – Hot Spot to visualize the heatmap
Methods
The APP works within an automatically or manually outlined tumor region of interest (ROI). Within each ROI, nuclei are detected and classified into Ki-67 positive or negative. The first image processing step involves a segmentation of all nuclei in the ROI. The HDAB-DAB color deconvolution band is used to detect positively stained nuclei and a multiplication of the red and blue color band is used to detect negative nuclei. A method for nucleus separation which is based on shape, size and nuclei probability is used, employing a fully automated watershed-based nucleus segmentation technique. As a post-processing step, nuclei areas that are too small are removed. The image obtained after post-processing is used to determine the output variables.
Staining Protocol
There is no staining protocol available.
Keywords
Cancer, neuroendocrine, Ki67, Ki-67, Proliferation index, Digital pathology, Image analysis, IHC
References
USERS
This APP was developed for Dr. Sönke Detlefsen, Department of Clinical Pathology, Odense University Hospital
LITERATURE
1. Scholzen, T., Gerdes, J. The Ki-67 protein: from the known and the unknown, Journal of Cellular Physiology 2000, 182 (3), 311-322, DOI.
2. Nadler, A. et. al. Ki-67 is a reliable pathological grading marker for neuroendocrine tumors, Virchows Archiv 2013, 462 (5), 501-505, DOI.