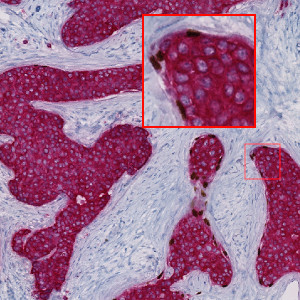
Image of a slide physically double stained with p63 (brown chromogen) and CK7/19 (red chromogen). Notice how only some structures display p63 positive nuclei.
#20101
Automated, accurate and objective
Manual exclusion of non-invasive tumor can in some specimens be challenging, even for an experienced pathologist. In cancers with a diffuse growth pattern, this can also be highly labor intensive.
With clinical breast cancer guidelines prescribing assessment of ER, PR, Ki-67, and HER2 on invasive cancers (ref. 1-3), the need for exclusion of noninvasive structures becomes essential. Biomarker analysis in hot spots is especially prone to bias by the non-invasive tumor.
The Invasive Tumor Detection APP automatically detects the invasive tumor. The algorithm integrates into a full biomarker analysis workflow, for automatic hot spot detection, accurate biomarker quantification, and visualization of tumor heterogeneity supporting the pathologist’s decisions.
Invasive tumor detection method
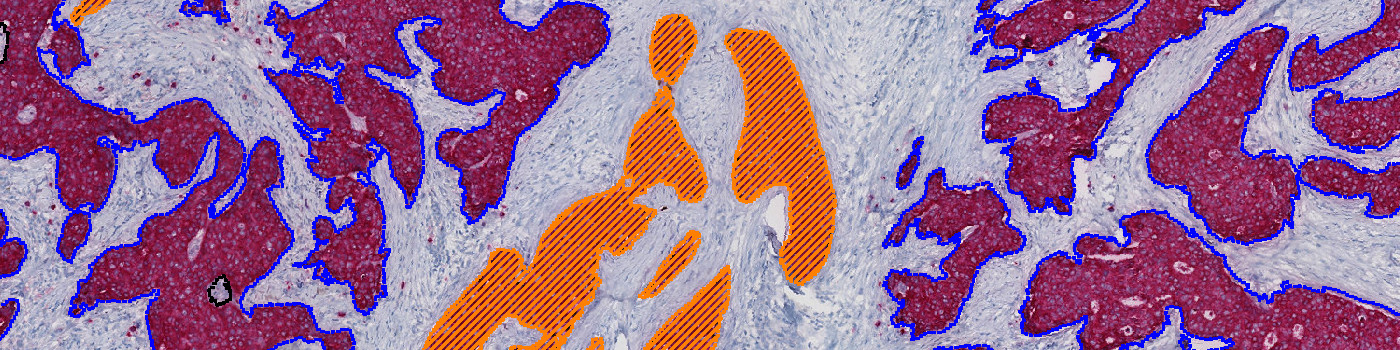
The Invasive Tumor Detection APP is based on a physical double stain of p63+CK7/19. Invasive tumor components are identified as regions positive for CK, whereas non-invasive tumor components are identified as regions positive for both CK and p63, and are excluded.
Biomarker assessment workflow
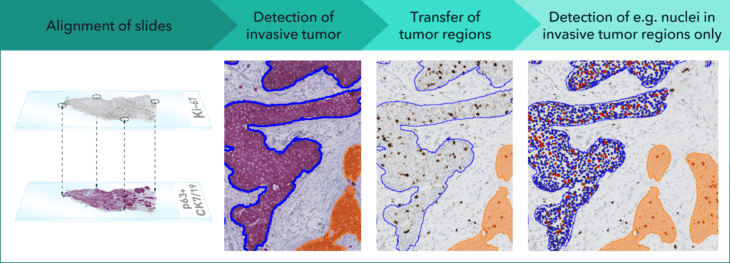
In combination with an analytical biomarker APP, the Invasive Tumor Detection APP provides an objective supplement to manual scoring. Results provided by both APPs must be confirmed by a qualified pathologist.
In the workflow illustrated above, serial sections of p63+CK7/19 and Ki-67 are virtually aligned. The p63+CK7/19 slide is evaluated for invasive and non-invasive tumor, which is transferred to the Ki-67 slide. Subsequent biomarker analysis within invasive tumor structures is accurate and according to clinical guidelines.
In EU/UK: CE IVD – for use in diagnostic procedures
Benefits of invasive tumor detection
Clinical studies
The clinical performance of the APP was determined by comparing non-invasive tumor structures as identified by the APP to non-invasive tumor structures as identified by a pathologist. Good agreement was obtained for all three sites, with mean DICE scores of 0.94, 0.81, and 0.91, respectively.
| Site | Samples (n) | Mean Dice Score | Lower 95% CI | Upper 95% CI |
|---|---|---|---|---|
| Odense University Hospital | 41 | 0.944 | 0.913 | 0.975 |
| Pathologist Diagnostics Jarutat | 39 | 0.810 | 0.731 | 0.888 |
| University Medical Center Groningen | 40 | 0.908 | 0.857 | 0.959 |
| Total | 120 | 0.888 | 0.854 | 0.922 |
Slide scanners: Hamamatsu NanoZoomer HT 2.0, Philips UltraFast 1.6
Stain vendors: p63: Roche Ventana/Zeta, CK7:Dako, CK19: AH-diagnostics/Cell Marque/Roche Ventana
CE-marked for in vitro diagnostics
The clinical performance of the APP was validated in collaboration with:
References
1. Assessment of Ki67 in Breast Cancer: Updated Recommendations From the International Ki67 in Breast Cancer Working Group, JNCI: Journal of the National Cancer Institute, Volume 113, Issue 7, July 2021, Pages 808–819
Torsten O Nielsen, MD, PhD, FRCPC, Samuel C. Y Leung, MSc, David L Rimm, MD, PhD, Andrew Dodson, MPhil, FIBMS, CSci, Balazs Acs, MD, PhD, Sunil Badve, MBBS, MD, FRCPath, Carsten Denkert, MD, Matthew J Ellis, MB, BChir, BSc, PhD, FRCP, Susan Fineberg, MD, Margaret Flowers, PhD, Hans H Kreipe, MD, Anne-Vibeke Laenkholm, MD, Hongchao Pan, PhD, Frédérique M Penault-Llorca, MD, PhD, Mei-Yin Polley, PhD, Roberto Salgado, MD, PhD, Ian E Smith, MD, FRCP, FRCPE, Tomoharu Sugie, MD, PhD, John M. S Bartlett, BSc, PhD, FRCPath, Lisa M McShane, PhD, Mitch Dowsett, BSc, PhD, Daniel F Hayes, MD,
2. Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline Focused Update. Arch Pathol Lab Med. 2018 Nov;142(11):1364-1382
Wolff AC, Hammond MEH, Allison KH, Harvey BE, Mangu PB, Bartlett JMS, Bilous M, Ellis IO, Fitzgibbons P, Hanna W, Jenkins RB, Press MF, Spears PA, Vance GH, Viale G, McShane LM, Dowsett M.
3. Estrogen and Progesterone Receptor Testing in Breast Cancer: ASCO/CAP Guideline Update. J Clin Oncol. 2020 Apr 20;38(12):1346-1366.
Allison KH, Hammond MEH, Dowsett M, McKernin SE, Carey LA, Fitzgibbons PL, Hayes DF, Lakhani SR, Chavez-MacGregor M, Perlmutter J, Perou CM, Regan MM, Rimm DL, Symmans WF, Torlakovic EE, Varella L, Viale G, Weisberg TF, McShane LM, Wolff AC.