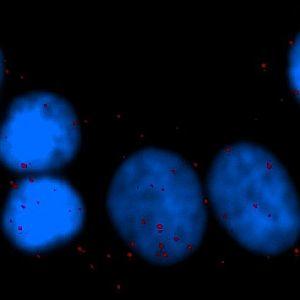
One field of view of the original cell line image (scaled down to fit this space).
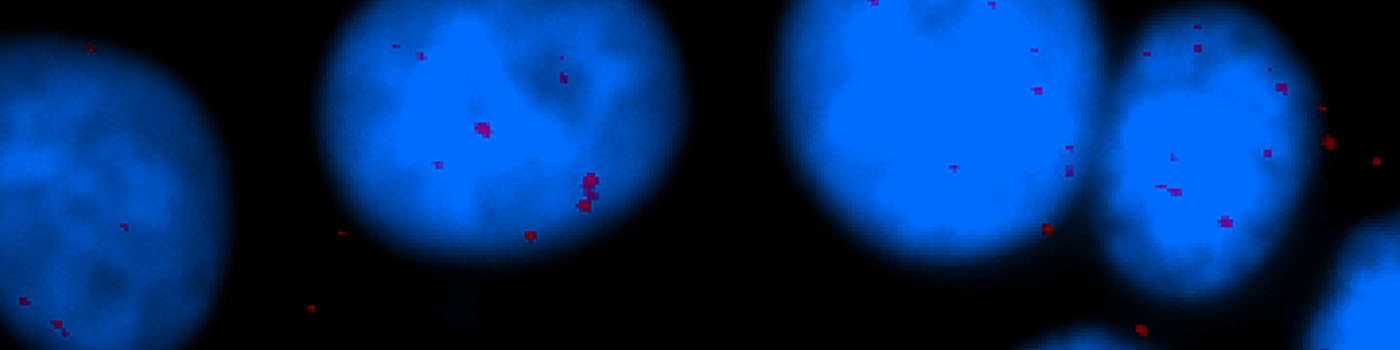
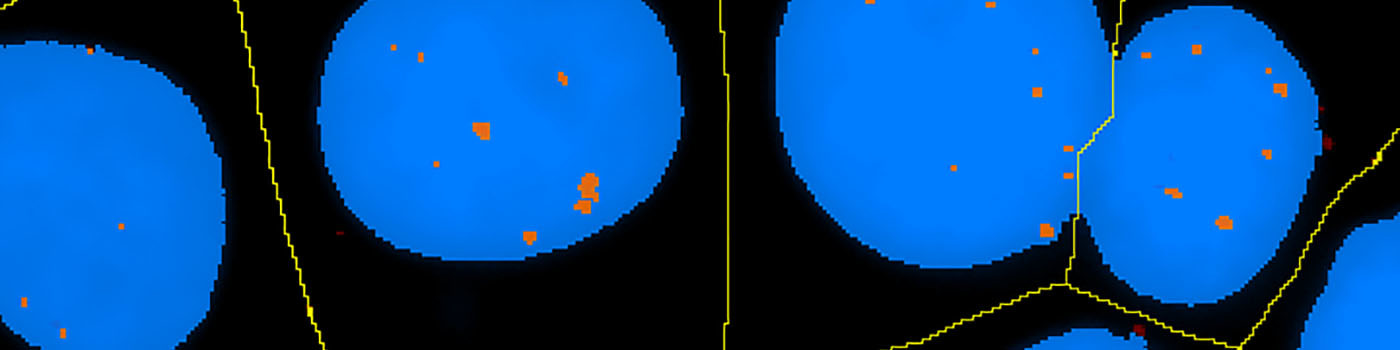
#10056
HER2 overexpression/amplification is linked with poor prognosis in early breast cancer. Co-expression of HER2 and HER3 is associated with endocrine and chemotherapy resistance, driven not simply by expression but by signaling via HER2:HER3 or HER2:HER2 dimers. Proximity ligation assays (PLAs) detect protein–protein complexes at a single-molecule level and allow study of signaling pathways in situ. PLA has been shown to successfully and reproducibly detect HER2:HER2 and HER2:HER3 protein complexes in vivo. It has also been shown that a significant association (P<0.00001) was identified between HER2 homodimerization and HER2 gene amplification, where high levels of HER2:HER2 dimers were significantly associated with reduced relapse free and overall survival.
Quantitative Output variables
Methods
The number of in situ PLA signals per nuclei was counted by automated image analysis. Fully automated nuclei delineation was initiated by identifying cell nuclei using the signal from the nuclear Hoechst staining (blue). Touching nuclei were separated using a combination of distance transformation and watershed segmentation. In situ PLA signals were counted by enhancing point-like signals and defining a true signal as a local intensity maximum above a background threshold.
Staining Protocol
Proximity ligation assay, see [2].
The slides are de-waxed and rehydrated. Antigens are retrieved by incubating the slides in 10 mM citrate buffer pH 6.0 at 96°C for 40 min in a waterbath.
The slides are washed twice with TBS (150 mM NaCl, 10 mM Tris pH 7.4) containing 500 µl 1 M glycine pH 8.5 and once with TBS for 5 min at room temperature, PLA was carried out using DUOLink™ kit (OLINK, Uppsala, Sweden) according to the manufacturer’s instructions.
Following blocking (DUOLink™ kit), the following antibodies were used overnight at 4°C:
HER2 ([1:200], DAKO, Cambridge, UK)
HER2 ([1:10] Novacastra, Newcastle upon Tyne, UK)
HER3 ([1:200] clone 2F12, Labvision, Cheshire, UK)
PLA plus and minus probes (containing the secondary antibodies conjugated with oligonucleotides) were added and incubated at 37°C for 2 h. Further oligonucleotides were added, to allow hybridization with PLA probes and ligase. The DNA was then amplified and detection carried out using the 563 detection kit (including Hoechst 33342 dye nuclear staining), resulting in red fluorescence signals. Finally, the slides are mounted with Vectashield mounting media (Vector Lab., Inc., Burlingame, CA, USA).
Keywords
Breast cancer, HER2, HER3, Proximity ligation assay
References
USERS
This APP was developed for, and validated by, Dr. Melanie Spears, Ontario Institute for Cancer Research (OICR).
LITERATURE
1. Hammond, M. E. H. et. al., American Society of Clinical Oncology/College of American Pathologists Guideline Recommendations for Immunohistochemical Testing of Estrogen and Progesterone Receptors in Breast Cancer, J. Clin Oncol 2010, 28 (16), 2784-95, DOI
2. Spears, M. et. al., In situ detection of HER2:HER2 and HER2:HER3 protein-protein interactions demonstrates prognostic significance in early breast cancer, Breast Cancer Res Treat 2012, 132 (2), 463-70, DOI
3. Jung, C. et. al., Segmenting Clustered Nuclei Using H-minima Transform-Based Marker Extraction and Contour Parameterization, IEEE Transactions on Biomedical Engineering 2010, 57 (10), 2600-2604, DOI
4. Abd El-Rehim, D. M. et. al., Expression and co-expression of the members of the epidermal growth factor receptor (EGFR) family in invasive breast carcinoma, Br J Cancer 2004, 91 (8), 1532-42, DOI
5. Bartlett, J. M. et. al., Type 1 receptor tyrosine kinase profiles identify patients with enhanced benefit from anthracyclines in the BR9601 adjuvant breast cancer chemotherapy trial, J. Clin Oncol 2008, 26 (31), 5027-35, DOI
6. Fredriksson, S. et. al., Protein detection using proximity-dependent DNA ligation assays, Nat. Biotechnol 2002, 20 (5), 473-77, DOI
7. Fredriksson, S. et. al., Multiplexing protein detection by proximity ligation for cancer biomarker validation, Nat. Methods 2007, 4 (4), 327-9, DOI
8. Soderberg, O. et. al., Direct observation of individual endogenous protein complexes in situ by proximity ligation, Nat. Methods 2006, 3 (12), 995-1000, DOI
9. Soderberg, O. et. al., Characterizing proteins and their interactions in cells and tissues using the in situ proximity ligation assay, Nat. Methods 2008, 45 (3), 227-32, DOI
10. Spears, M. et. al., Type 1 receptor tyrosine kinases as predictive or prognostic markers in early breast cancer, Biomarker Med. 2008, 2 (4), 397-407, DOI
11. Tovey, S. M. et. al., Outcome and human epidermal growth factor receptor (HER) 1–4 status in invasive breast carcinomas with proliferation indices evaluated by bromodeoxyuridine labeling, Breast Cancer Res 2004, 6 (3), R246-51, DOI
12. Tovey, D. M. et. al., Can molecular markers predict when to implement treatment with aromatase inhibitors in invasive breast cancer?, Clin Cancer Res 2005, 11 (13), 4835-42, DOI
13. Witton, C. J. et. al., Expression of the HER1–4 family of receptor tyrosine kinases in breast cancer, J Pathol 2003, 200 (3), 290-97, DOI