
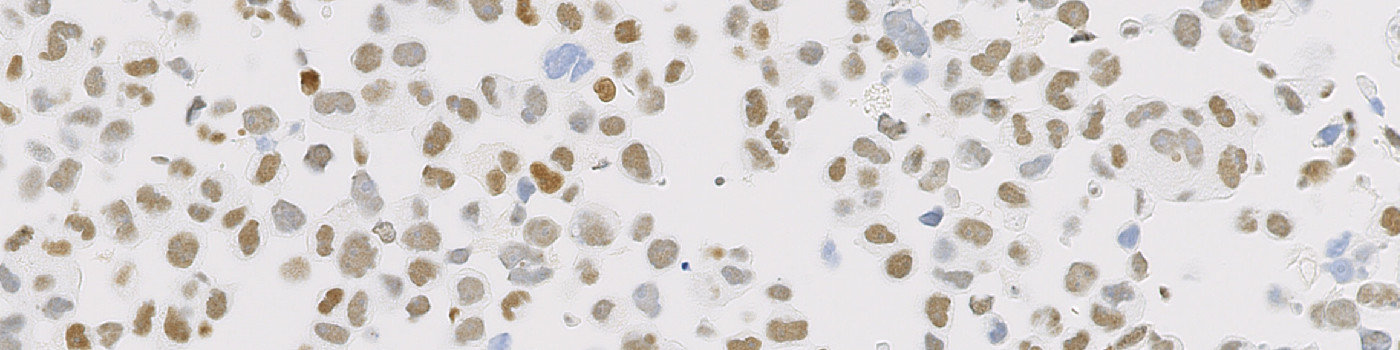
Raw image of cell line cores with different ER expression.
#10102
The availability of genetically defined reference materials, offers an industry standard for development and quality control of IHC assays, directly, thereby improving the accuracy and reproducibility. Using engineered cell lines as quality control material to assess the performance of IHC assays eliminates the variability associated with patient-derived reference standards.
This APP can be used for quality control of ER strong positive, intermediate positive and negative cell line material to ensure that each cell line block complies with the expected ER expression before the material is used as reference standard for IHC assays. The APP quantifies the ER expression in each cell line core and relates this expression to a known reference value for each cell line expression level.
Quantitative Output variables
The output variables obtained from this protocol are:
Methods
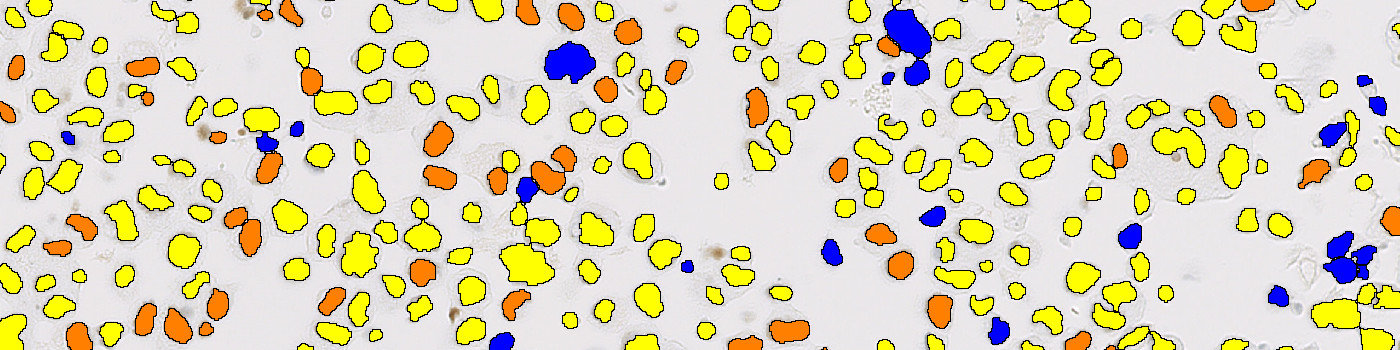
The first image processing step involves a segmentation of all nuclei in the ROI. This is done by using an HDAB-DAB color deconvolution band to detect positively stained nuclei and the Haematoxylin-HDAB color deconvolution band to detect negative nuclei. A method for nuclei separation which is based on shape, size and nuclei probability is used, employing a fully automated watershed-based nuclei segmentation technique. Then the positive nuclei are subdivided into three categories based on staining intensities. As a post-processing step, nuclei areas that are too small are removed. The image obtained after post-processing (see FIGURE 3), is used for quantifying the ER expression in each cell line core based on the H-score. As a final step the H-score is related to a known reference value for each cell line expression level.
Staining Protocol
There is no staining protocol available.
Keywords
Estrogen receptor, cell line, quality control, reference standards, image analysis
References
USERS
The APP was developed for Horizon cell lines.
LITERATURE
There are currently no references.