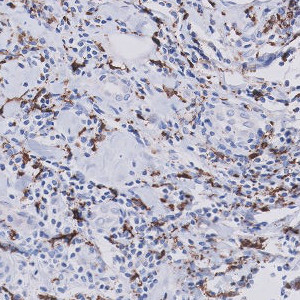
Breast tissue with CD68 positive nuclei.
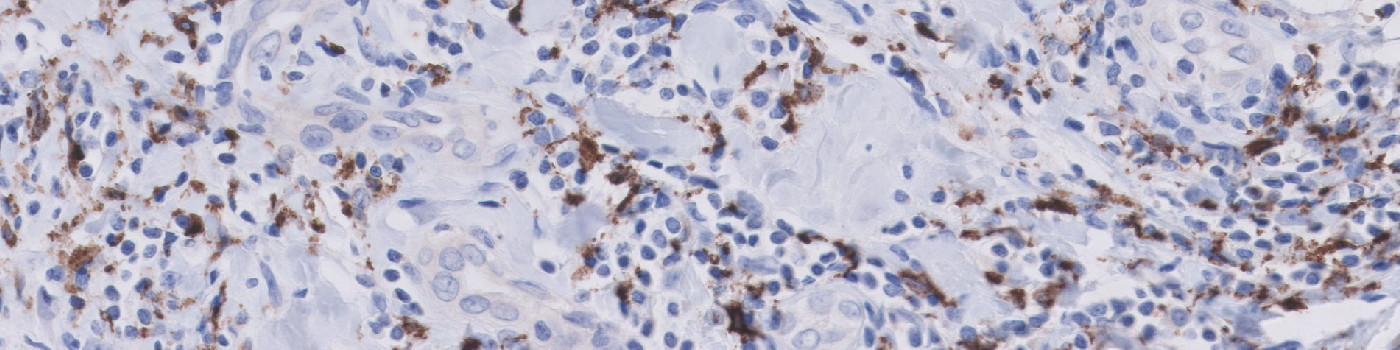
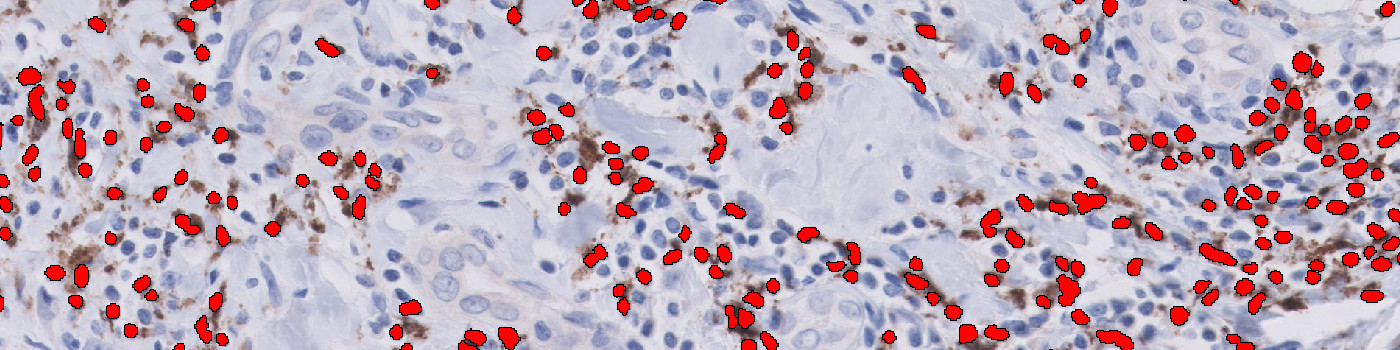
#10129
Breast cancer development and progression could be affected by its tumour-associated macrophages (TAMs). More specifically, a disbalance between TAM subtypes: M1-like and M2-like macrophages might lead to unfavourable outcome.
The present APP works on serial section TMAs stained with CD68 and CD163. The APP quantifies the number of positive cells for both stains, which enables the calculation of the M2/M1 ratio as #CD163/(#CD68 – #CD163).
Auxiliary APPs
APP1: “01 CD68 quantification”
APP2: “02 CD163 quantification”
Quantitative Output variables
The output variables obtained from this protocol are:
From which #M1 can be calculated as:
Workflow
Step 1: De-array the CD68 and CD163 TMAs
Step 2: Load and run the APP “01 CD68 quantification” as a batch process on the CD68 cores
Step 3: Load and run the APP “02 CD163 quantification” as a batch process on the CD163 cores
Methods
First, a pre-processing step enhances the nuclear stain by hematoxylin color deconvolution. Then a blob filter is applied to identify the individual nuclei. During post-processing the individual nuclei are classified as negative or positive based on the amount of surrounding CD68 and CD163 stain. Also, the size of the nuclei is taken into account to exclude tumor cells.
Staining Protocol
There is no staining protocol available.
Keywords
CD68, CD163, Breast cancer, Digital pathology, TMA, Image analysis, Cell quantification, TME
References
USERS
This APP was developed for Dr. Bert Van Der Vegt, University Medical Center Groningen, Netherlands.
LITERATURE
There are currently no references.