Endometrial tissue section. Overview image at 0.5x.
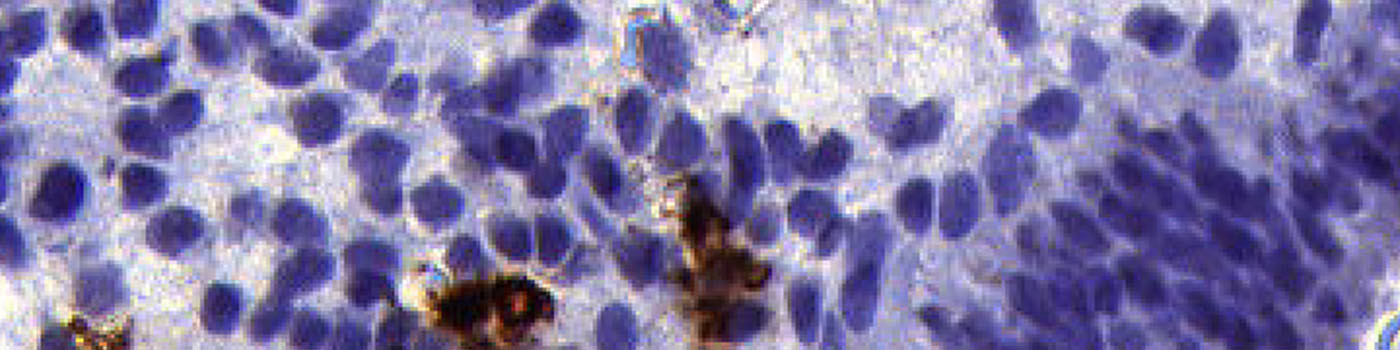
#10042
Uterine natural killer (uNK) cells in the pre-pregnancy endometrium have been associated with recurrent miscarriage. Immunohistochemical (IHC) analysis of endometrial tissue sections has demonstrated increased density of CD56+ CD16- uNK cells in the mid-luteal phase endometrium in women suffering from idiopathic recurrent miscarriage and recurrent implantation failure compared to fertile controls, see [1].
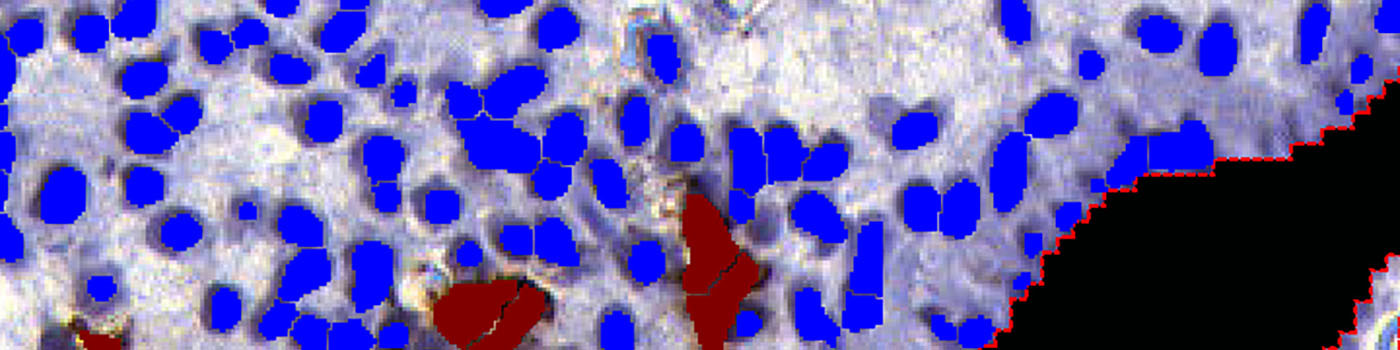
This APP measures the ratio of uNK cells among the stromal cells near the epithelial edge. Cells in the glands and in the epithelial edge itself do not contribute to the calculations.
This APP is designed to work on approx. 3 µm sections of endometrial tissue immunostained with anti-CD56 monoclonal antibody and counterstained with Mayer’s haematoxylin.
Auxiliary APPs
Auxiliary APPs are used for additional process steps, e.g. finding Region of Interest (ROI).
IDENTIFY TISSUE
The auxiliary APP ’01 Find Tissue’ can be used to automatically detect the region of interest (ROI). The APP utilizes a Bayesian classifier to detect the tissue areas of interest (see FIGURE 1 and 2). It might, sometimes be needed to manually remove areas of no interest subsequently to e.g. exclude normal and necrotic tissue.
IDENTIFY TISSUE COMPONENTS
The auxiliary APP ’02 Find Epitheleal Edge’ can be used to automatically identify the epithelial edge within the tissue based on saturation and local shape. It outlines a final ROI up to 300 µm into the tissue from the epithelial edge, still excluding glands.
Quantitative Output variables
The output variables obtained from this protocol include:
Workflow
Step 1: Load auxiliary APP for tissue detection ’01 Find Tissue’ and run on the image to process. This outlines the tissue and the glands with a Region Of Interest (ROI) and saves time in the subsequent analysis steps. It may be necesarry to refine the ROI to remove e.g. glands or other residuals.
Step 2: Load auxiliary APP ’02 Find Epitheleal Edge’ and run on the image to identify the epithelial edge within the tissue based on saturation and local shape. It outlines a final ROI up to 300 µm into the tissue from the epithelial edge, still excluding glands.
Step 3: Load APP quantification protocol ’03 Count Cells’ and run on the images to process. The third image processing step identifies the uNK and stromal cells, within the specified ROI, and calculates the output variables.
Methods
This APP includes 3 separate steps, each step contributing to the final output.
The first step identifies the tissue and the glands, based on intensity and local shape, see [2]. The tissue is subsequently defined as the initial region of interest (initial ROI), and the glands are excluded from this region (see FIGURE 1 and 2). Manual interaction may be needed to refine the initial ROI e.g. removing additional glands.
The second step identifies the epithelial edge within the tissue based on saturation and local shape. It outlines a final ROI up to 300 µm into the tissue from the epithelial edge, still excluding glands (see FIGURE 1, 2, 3, 4).
The third step identifies uNK cells and stromal cells within the final ROI, and calculates the relevant output variables, including the ratio of uterine Natural Killer (uNK) cells to stromal cells (see FIGURE 5 and 6).
Staining Protocol
There is no staining protocol available.
Keywords
Endometrium, uterus, CD56, uterine Natural Killer cells, uNK, epithelial edge, haematoxylin, immunohistochemistry, quantitative, digital pathology, image analysis.
References