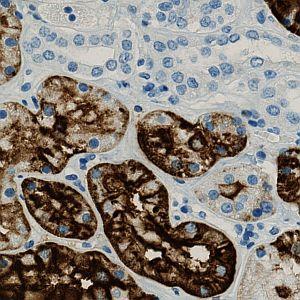
Original image.
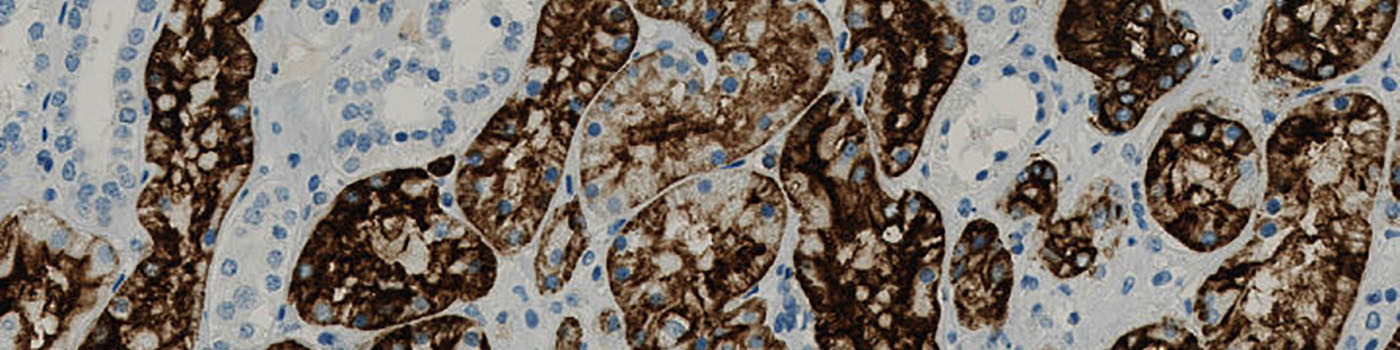
#10053
The purpose of the APP is to determine the number (using an unbiased counting frame), the area and the area fraction of the positive cells (positive cells are here: blue nuclei surrounded by brown cytoplasm). The APP can furthermore determine the extent and intensity of CD15 staining (brown tissue) based on optimized protocols on Benchmark Ultra (Ventana) and Autostainer (Dako) with various antibodies in order to quantify staining reactions, primarily in renal tumors for comparison of antibodies and platforms.
Regarding diagnostic application CD15 may be used for the subtyping of renal cell carcinoma and for the differential diagnosis of carcinomas vs. malignant mesothelioma and mesenchymal tumors. The ratio of CD15 positive tissue is an indicator for the progression and aggression of the disease.
In some neoplastic diseases like Hodgkin disease, myeloid leukaemias and gliomas, CD15 expression is inversely correlated to dedifferentiation and progression.
CD15 is a carbohydrate, also known as 3-fucosyl-N-acetyl-lactosamine, Lewis antigen X (LeX) and Leu-M1, a haemopoietic differentiation antigen with poorly understood functions which includes mediating phagocytosis and chemotaxis.
CD15 is expressed on most terminally differentiated myeloid cells (granulocytes, monocytes/histiocytes and Langerhans cells), while myeloid progenitor cells are rarely positive. The vast majority of lymphocytes are negative, but activated T-lymphocytes may become positive. CD15 is also found in epithelia of various organs such as breast (secretory epithelium), kidney (proximal tubules and loop of Henle), lung, urothelium, oesophagus and gastrointestinal tract. In the nervous system CD15 is present in astrocytes and variably in oligodendrocytes and neurons.
In CD15 positive cells the antigen expression may be membranous, diffuse cytoplasmic or related to the Golgi zone.
Among haematolymphoid neoplasms CD15 is expressed in Reed-Sternberg and Hodgkin cells in classical Hodgkin lymphoma in most cases (up to ~90%) while the neoplastic L&H cells in nodular lymphocyte predominant Hodgkin lymphoma only express CD15 in a small percentage of cases. Non-Hodgkin lymphomas are usually CD15 negative, though small proportions (10-15%) of CD15 positive cases have been found. These cases are particularly among peripheral and angioimmunoblastic T-cell lymphomas, mycosis fungoides and diffuse large B-cell lymphoma. Acute lymphoblastic leukaemia is rarely CD15 positive. In acute myeloid leukaemia, CD15 positive cells occur as a sign of maturation. Chronic myeloid leukaemia is regularly CD15 positive, the cells reacting in a heterogeneous manner.
CD15 can also be demonstrated in a high proportion of adenocarcinomas from, e.g., breast, lung, stomach, colon, pancreas, biliary tract, ovary, urinary tract and skin. Hepatocellular carcinoma, endocrine neoplasms, squamous cell carcinoma and urothelial carcinomas are mostly CD15 negative. Renal cell tumors, clear cell and papillary carcinomas express CD15 in the majority of cases, while oncocytoma and chromophobe carcinoma more seldom express this antigen.
Malignant mesothelioma is virtually always CD15 negative, but a small proportion of particularly desmoplastic mesothelioma is reported positive. Gliomas show little CD15 expression. Among germ cell tumors, only teratoma is reported positive. Mesenchymal tumors and malignant melanoma are virtually always negative.
Several monoclonal antibodies are commercially available of which clone Carb-3 seems to be the most sensitive. Clone MMA may also give good results, while clone BY87 appears less sensitive. For all clones, efficient heat induced epitope retrieval (HIER) is mandatory, an alkaline buffer is recommended.
Normal kidney is primary choice for control tissue: The proximal tubules should be strongly stained while the distal tubules remain unstained.
Quantitative Output variables
Methods
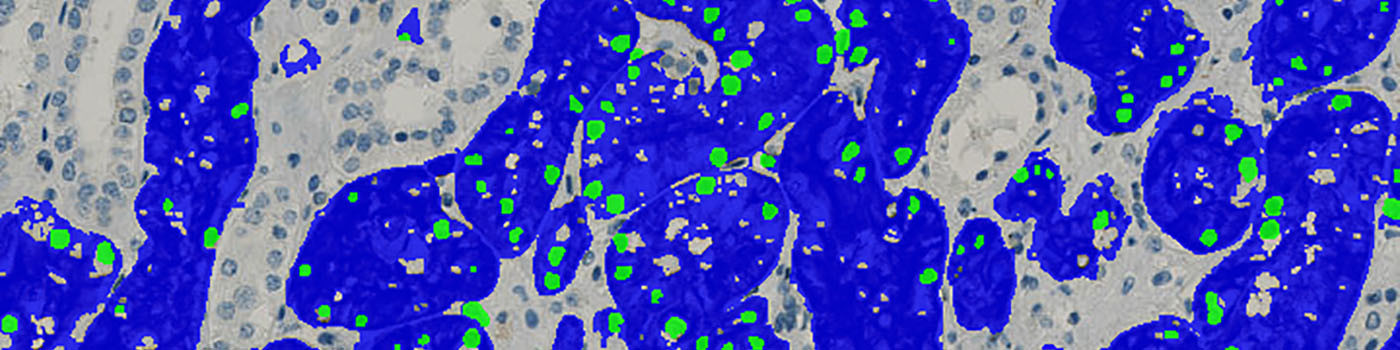
The first processing step in the APP is the nuclei and tissue detection, which involves segmentation of all blue nuclei and CD15 (brown) tissue within the ROI (which is drawn by the user) (see FIGURE 2). The tissue is identified by assigning a label probability to all pixels in the image, resulting in a label probability image. The label probability image is found by an intensity dictionary – a dictionary with small image patches. The intensity dictionary can be coupled to a label dictionary from which the label probability image is obtained. Based on this image, segmentation of the tissue can be done by choosing the most probable label in each pixel, see [1]. A method for nuclei separation which is based on shape, size and nuclei probability is used, employing a fully automated watershed-based nuclei segmentation technique. The method is an extension of the method proposed by Jung and Kim, see [2], where an h-minima transform is used before applying the watershed. This is only applied to the nuclei and not the CD15 (brown) tissue.
Thereafter, the APP removes all nuclei that are not considered positive (i.e. within CD15, brown, tissue) by first observing the nearby tissue (see FIGURE 3) and secondly observing the shape of the nuclei (see FIGURE 4).
Finally, tissue fractions below a certain area size is removed.
NOTE: The APP requires a manually drawn ROI around the tissue of interest.
Staining Protocol
There is no staining protocol available.
Additional information
This APP was developed in cooperation with Professor Mogens Vyberg from NordiQC and Aalborg University Hospital.
Keywords
Kidney, CD15, expression, intensity, tumor, cancer, number, area
References
USERS
This APP was developed in cooperation with Professor Mogens Vyberg from NordiQC and Aalborg University Hospital.
LITERATURE
1. Dahl, A.L. et. al., Learning Dictionaries of Discriminative Image Patches, British Machine Vision Conference 2011, DOI
2. Jung, C. et. al. Segmenting Clustered Nuclei Using H-minima Transform-Based Marker Extraction and Contour Parameterization, IEEE Transactions on Biomedical Engineering 2010, 57 (10), 2600-2604, DOI