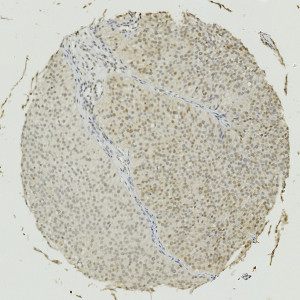
Image of a TMA core with bladder tumor tissue stained by IHC for Annexin 10.

#10012
The annexin family is composed of 12 eukaryotic members participating in diverse important biological and physiological processes including anti-coagulation, endocytosis, exocytosis, immune suppression, differentiation, and tissue growth see, [1][2][3][4]. Many studies have shown the annexins to be among the genes consistently differentially expressed in neoplasia. Increasing evidence also indicates that cancer-specific changes in annexin expression or in their subcellular localization contributes to development and progression of cancer by affecting cell signaling pathways, cell motility, tumor invasion and metastasis, angiogenesis, apoptosis, and drug resistance, see [4][5]. Furthermore, Annexin 10 has been reported to be correlated with poor prognosis in hepatocellular carcinoma [6], gastric carcinoma [7], and latest in urothelial carcinoma, see [8].
This APP can be used for quantifying the nuclear expression of Annexin 10. The approach to calculating expression used here, was demonstrated to have a high correlation to manual scoring in a study involving 300 patients (R2 = 0.70) (publication pending).
Quantitative Output variables
The output variables obtained from this protocol include:
Methods
The method used for computing the Annexin 10 expression is started by detecting nuclei using a novel pattern recognition method adapted from Dahl et al , see [8], which is followed by a step that separates adjacent nuclei, see [9]. The detected nuclei are classified as either positive or negative based on a computation of DAB intensity, obtained using color de-convolution.
This leads to the calculation of the positive area fraction, as the area of positive nuclei divided by the total area of nuclei (within the outlined ROI). This is followed by the measurement of the average DAB intensity across all positive nuclei, which is subtracted from 255, to associate higher values with a high staining intensity, which is more intuitive. The expression is then calculated by multiplying the intensity with the area fraction.
The actual implementation of the pattern recognition allows the user to choose settings for nuclear detection sensitivity and classification into positive and negative, in order to account for differences in local staining protocols.
It should be noted that this approach to quantifying biomarker expression is vulnerable to variations in protocols for tissue preparation (fixation time, tissue thickness, reagents, auto-stainer type and settings etc.). Using this APP for TMA study designs, however, is robust for the cores within the TMA.
This APP requires manual outlining of tumor regions.
Keywords
Annexin 10, bladder cancer, metastasis, immunohistochemistry, image analysis, quantitative, digital pathology.
References
USERS
This APP was developed for, and validated by, Niels Fristrup MD and Dr. Lars Dyrskjøt Andersen, Center for Molecular Clinical Cancer Research Department of Molecular Medicine (MOMA) Aarhus University Hospital.
LITERATURE
1. Gerke, V., et. al. Annexins: from structure to function, Physiol Rev 2002, 82 (2), 331-71, DOI
2. Hayes, M.K., et. al. Annexins and disease, Biochem Biophys Res Commun 2004, 322 (4), 1166-1170, DOI
3. Gerke, V., et. al. Annexins: linking Ca2+ signalling to membrane dynamics, Nat Rev Mol Cell Biol 2005, 6 (6), 449-61, DOI
4. Hayes, M.K., et. al. Annexinopathies, Subcell Biochem 2007, 45, 1-28, DOI
5. Mussunoor, S., et. al. The role of annexins in tumour development and progression, J Pathol 2008, 216 (2), 131-40, DOI
6. Liu, S.H., et. al. Down-regulation of annexin A10 in hepatocellular carcinoma is associated with vascular invasion, early recurrence, and poor prognosis in synergy with p53 mutation, Am J Pathol 2002, 160 (5), 1831-7, DOI
7. Kim, J., et. al. Reduced expression and homozygous deletion of annexin A10 in gastric carcinoma, Int J Cancer 2009, 125 (8), 1842-50, DOI
8. Dahl, A.L., et. al. Learning Dictionaries of Discriminative Image Patches, British Machine Vision Conference 2011, DOI
9. Jung, C., et. al. Segmenting Clustered Nuclei Using H-minima Transform-Based Marker Extraction and Contour Parameterization, IEEE Transactions on Biomedical Engineering 2010, 57 (10), 2600-4, DOI