
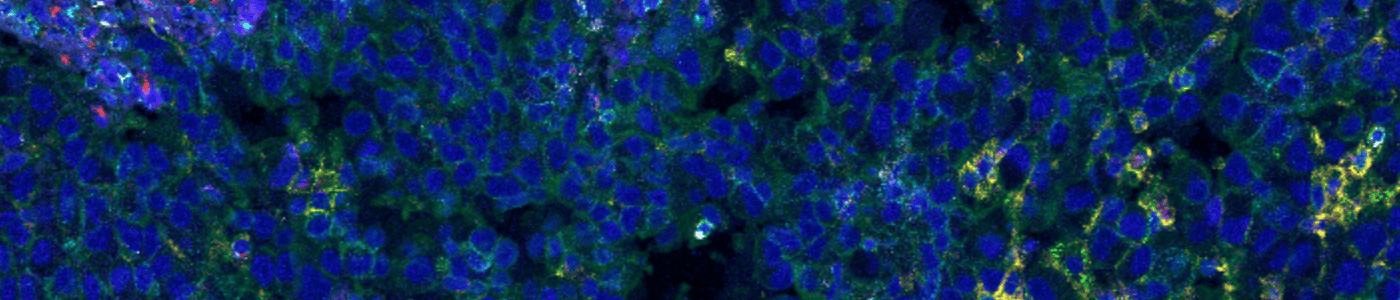
Multiplex IMC image before running this APP.
#10184
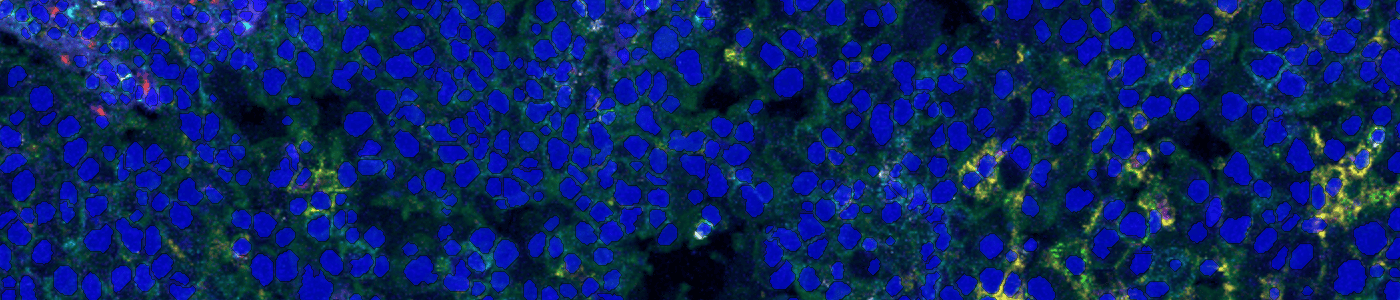
Automatically segment cells in your IMC (imaging mass cytometry) images to perform multiplexed nuclear profiling. All you need are images acquired from your Hyperion XTi Imaging system. This APP runs on the DNA channels, ensuring that nuclei are segmented accurately.
After analysis is complete, define phenotypes with the Phenoplex™ Guided Workflow and explore your results with the Data Exploration tools (available on Discovery and Phenoplex™).
As with all of our Quickstart APPs and custom-build APPs, your image analysis can be scaled by queueing APPs to run in the background on an unlimited number of images and cores. This is possible with the “batch analysis” feature available on both Discovery and Phenoplex™.
Working with multimodal data sets or TMAs? No problem. Run this APP after using the Tissuealign™ feature to align your images at the cellular level, or the Tissuearray™ feature to de-array your tissue cores.
Quantitative Output variables
Workflow